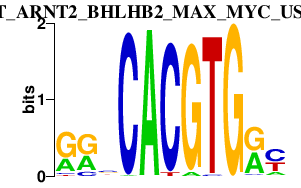
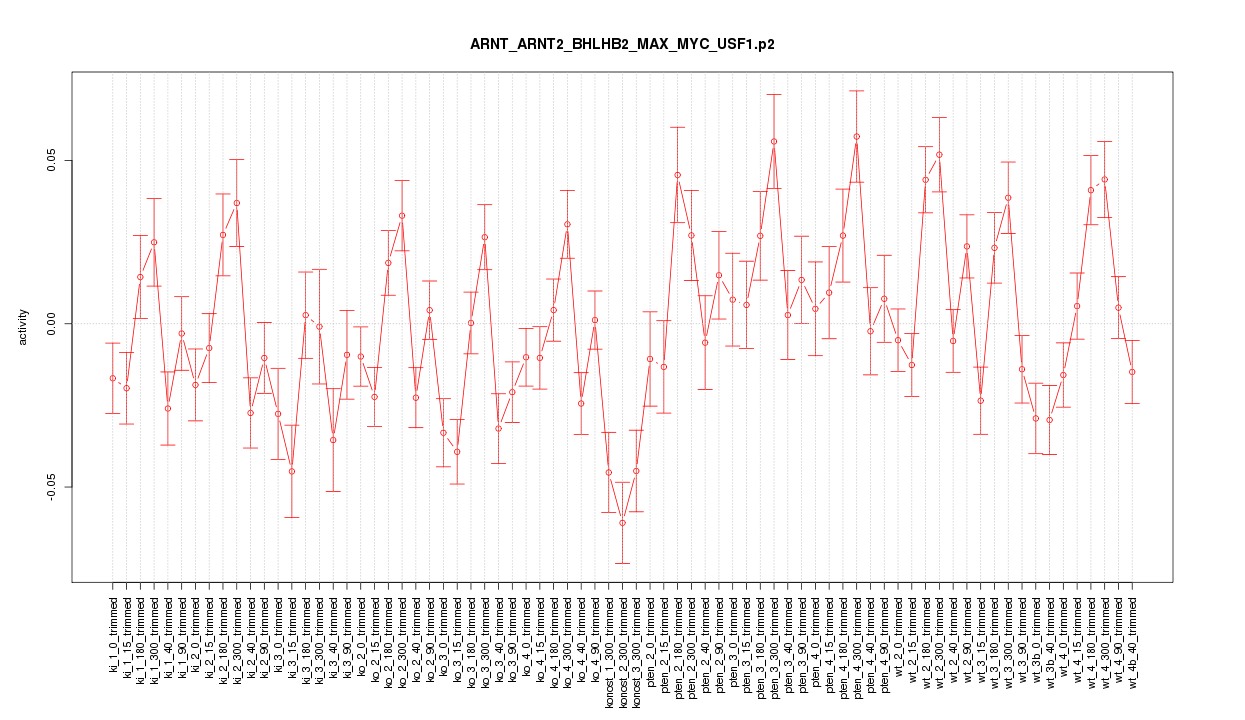
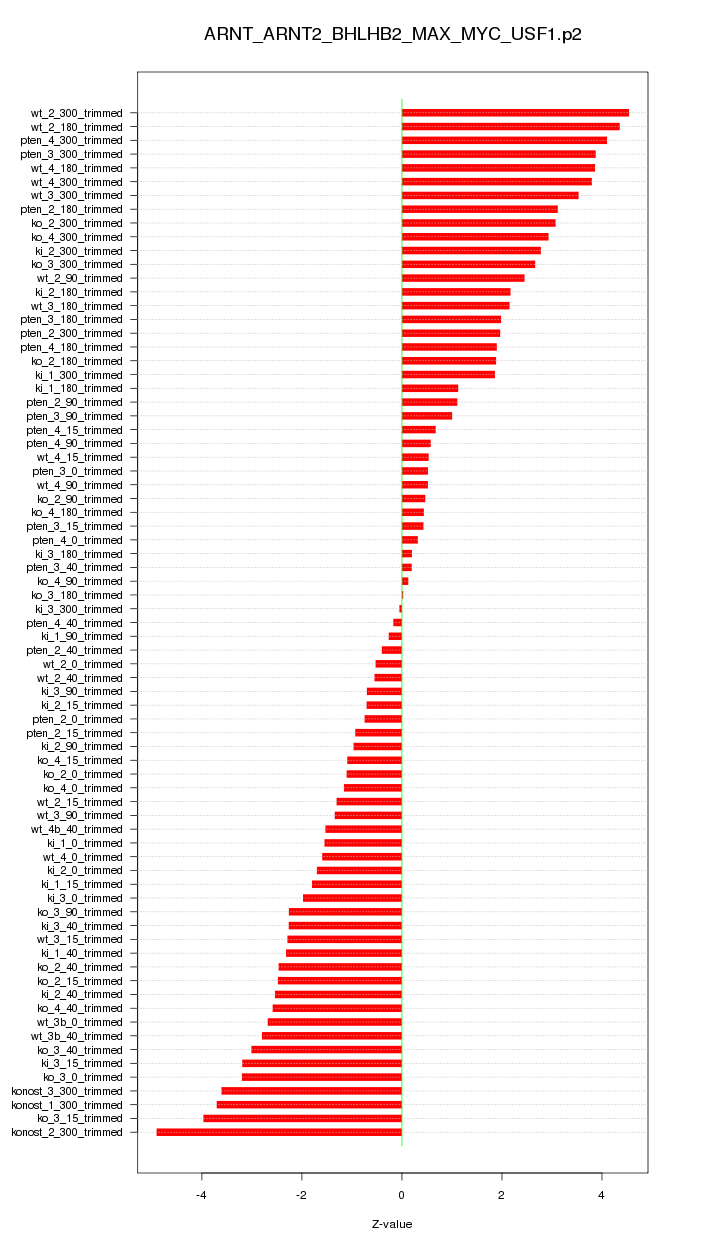
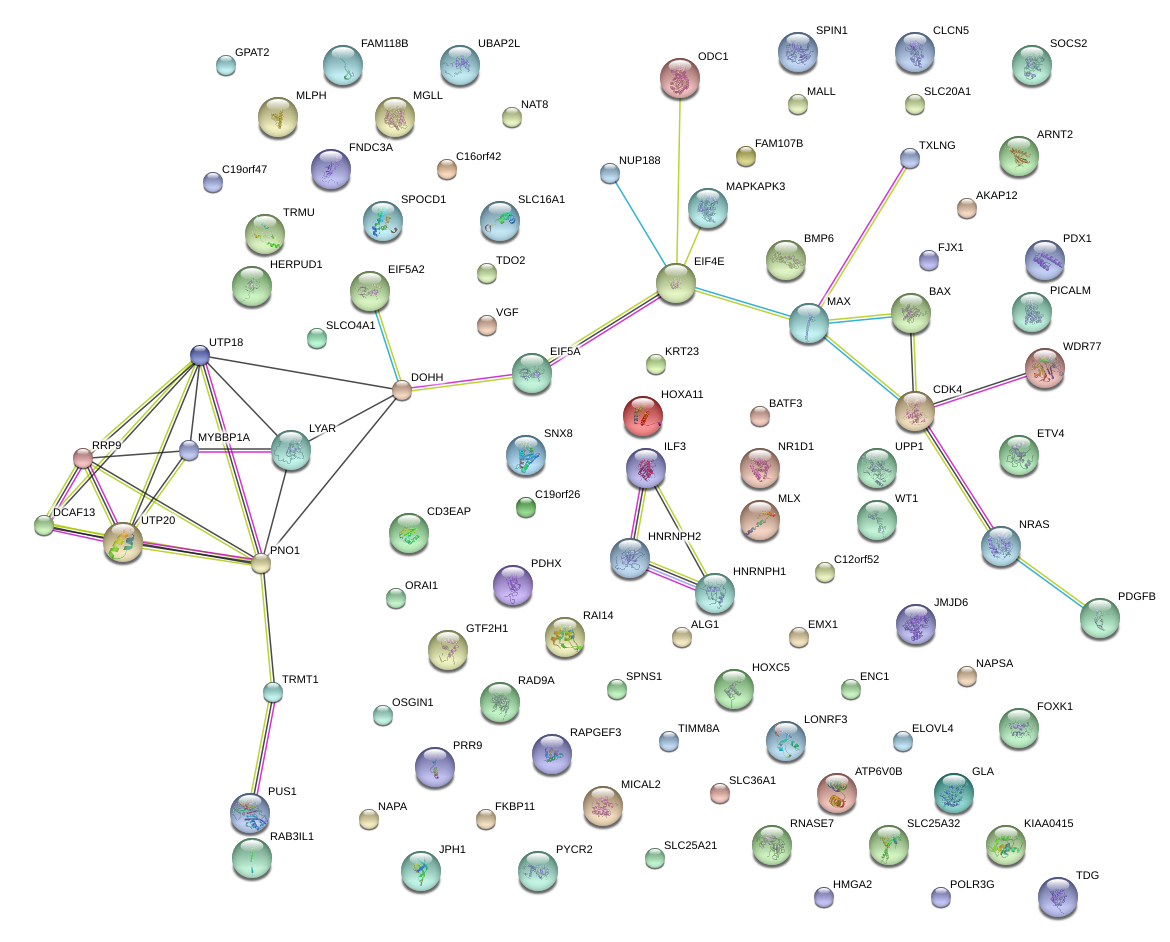
|
chr11_-_61684981
|
28.403
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr3_-_170626425
|
21.240
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr6_+_151561094
|
21.009
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr2_+_113403433
|
19.956
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr1_-_212873306
|
19.403
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr3_-_170626381
|
17.879
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr1_-_212873197
|
17.841
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr1_-_212873104
|
16.920
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chrX_+_118108575
|
16.881
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chrX_+_118108595
|
15.938
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr4_-_99850280
|
15.530
|
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr6_+_151646634
|
14.907
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_+_45909466
|
14.578
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr13_+_28494136
|
14.553
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr4_-_99850242
|
14.027
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chrX_-_100603673
|
11.593
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr1_-_53164014
|
11.228
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr1_-_32281577
|
11.043
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chrX_-_100603956
|
10.877
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr6_+_7726268
|
10.553
|
|
BMP6
|
bone morphogenetic protein 6
|
|
chr11_-_85780088
|
10.398
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_+_154193294
|
10.387
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr2_+_68384986
|
10.344
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr19_+_49458031
|
10.303
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr19_-_3500620
|
10.274
|
NM_031304
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase
|
|
chr11_-_85780105
|
10.227
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_58146108
|
10.097
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_58146110
|
10.092
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr9_+_131709991
|
10.075
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr12_+_66218141
|
9.927
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr12_-_58146025
|
9.804
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr7_-_27224762
|
9.708
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr11_+_35639734
|
9.605
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr1_+_44440642
|
9.484
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr19_-_48018277
|
9.342
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_-_53163985
|
9.309
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr12_-_49318663
|
9.296
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chrX_+_100663213
|
9.253
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr4_-_4291889
|
9.202
|
NM_017816
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr4_-_4291817
|
9.083
|
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chrX_+_100663205
|
9.078
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr13_+_49684388
|
9.044
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr12_+_54426831
|
9.019
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr8_-_104427373
|
8.926
|
NM_030780
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chrX_+_49832214
|
8.904
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr4_-_4291731
|
8.875
|
NM_001145725
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr9_+_131709970
|
8.772
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr19_+_10764896
|
8.697
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr12_-_48152860
|
8.647
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr19_+_45909868
|
8.607
|
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chrX_+_100663229
|
8.580
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr11_+_18344113
|
8.540
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr1_+_44440595
|
8.509
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr22_-_39640956
|
8.458
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr17_+_49337892
|
8.417
|
NM_016001
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr1_+_44440623
|
8.389
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_+_49337882
|
8.331
|
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr12_+_113623364
|
8.318
|
NM_032848
|
C12orf52
|
chromosome 12 open reading frame 52
|
|
chr11_-_85779821
|
8.291
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr19_-_40854283
|
8.234
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr17_+_40719064
|
8.224
|
NM_170607
NM_198204
NM_198205
|
MLX
|
MAX-like protein X
|
|
chr16_-_1401765
|
8.212
|
NM_001001410
|
C16orf42
|
chromosome 16 open reading frame 42
|
|
chr8_-_104427268
|
8.109
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr20_+_61273786
|
8.103
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr17_+_49337909
|
7.903
|
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast)
|
|
chr1_+_44440673
|
7.891
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr2_-_96700662
|
7.848
|
NM_207328
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr2_-_10588451
|
7.748
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr16_+_83986826
|
7.723
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr12_-_58146118
|
7.717
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_-_74722666
|
7.653
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr8_-_75233461
|
7.627
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr17_-_38256548
|
7.605
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr19_-_1237840
|
7.554
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr1_-_53164023
|
7.535
|
NM_023077
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr16_+_5121809
|
7.535
|
NM_019109
|
ALG1
|
asparagine-linked glycosylation 1, beta-1,4-mannosyltransferase homolog (S. cerevisiae)
|
|
chr11_+_67159421
|
7.524
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr5_+_150827138
|
7.500
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr5_+_150827204
|
7.495
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr6_-_80656870
|
7.453
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr12_+_122064448
|
7.435
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr17_-_74722629
|
7.428
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr11_+_12132063
|
7.425
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr5_+_34656595
|
7.391
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr8_-_13133956
|
7.341
|
|
|
|
|
chr2_-_110873353
|
7.325
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr11_+_126081618
|
7.265
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr12_+_132413776
|
7.250
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr17_-_4458618
|
7.242
|
NM_001105538
NM_014520
|
MYBBP1A
|
MYB binding protein (P160) 1a
|
|
chr12_+_104359557
|
7.189
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr1_-_115259381
|
7.185
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr2_+_238395863
|
7.155
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr2_-_110873638
|
7.143
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr5_-_73937248
|
7.086
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr1_-_113498828
|
7.064
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr7_+_48128738
|
7.011
|
NM_003364
|
UPP1
|
uridine phosphorylase 1
|
|
chr17_-_39093671
|
7.006
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr7_-_100808832
|
7.000
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr5_+_150827187
|
6.923
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr16_+_56965747
|
6.912
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr7_-_2354050
|
6.895
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr11_+_18343794
|
6.884
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr11_-_32457059
|
6.876
|
NM_000378
NM_024424
NM_024426
|
WT1
|
Wilms tumor 1
|
|
chr17_-_38256905
|
6.843
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr12_+_93964801
|
6.837
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr2_-_110873533
|
6.820
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr19_-_40854410
|
6.802
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr20_+_61273823
|
6.779
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr6_-_33385814
|
6.734
|
|
|
|
|
chr19_-_13227212
|
6.645
|
NM_001142554
NM_017722
NM_001136035
|
TRMT1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr17_-_41623652
|
6.616
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr5_+_89770680
|
6.595
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr16_+_28986024
|
6.594
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr12_+_101673895
|
6.571
|
NM_014503
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast)
|
|
chr19_+_10765106
|
6.554
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr2_-_110874142
|
6.483
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr10_-_14646237
|
6.456
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr8_+_104427612
|
6.440
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr1_-_226111977
|
6.391
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr8_+_104427659
|
6.382
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chrX_+_16804531
|
6.327
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr1_-_113498615
|
6.313
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chrX_-_100662905
|
6.230
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr14_+_21510384
|
6.190
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr16_+_56966025
|
6.188
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr3_-_51975879
|
6.173
|
NM_004704
|
RRP9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast)
|
|
chr17_-_74722517
|
6.159
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr22_-_39636906
|
6.155
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr3_+_50654558
|
6.151
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr19_+_49458129
|
6.134
|
|
BAX
|
BCL2-associated X protein
|
|
chr5_+_34656482
|
6.118
|
|
RAI14
|
retinoic acid induced 14
|
|
chr19_-_48018317
|
6.109
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_-_111991790
|
6.107
|
NM_024102
|
WDR77
|
WD repeat domain 77
|
|
chr14_-_65569221
|
6.098
|
NM_002382
NM_145112
NM_145113
NM_145114
NM_145116
NM_197957
|
MAX
|
MYC associated factor X
|
|
chr1_+_154193757
|
6.046
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr1_+_153190059
|
6.020
|
NM_001195571
|
PRR9
|
proline rich 9
|
|
chr7_+_4815258
|
5.986
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr5_+_34656367
|
5.963
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr3_-_127541160
|
5.958
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr9_-_140009169
|
5.932
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr20_-_36758362
|
5.922
|
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr17_-_74722843
|
5.872
|
NM_001081461
NM_015167
|
JMJD6
|
jumonji domain containing 6
|
|
chr12_+_57984992
|
5.817
|
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr7_-_2354010
|
5.812
|
|
SNX8
|
sorting nexin 8
|
|
chr8_-_134584012
|
5.803
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr9_-_140009194
|
5.787
|
NM_013379
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr4_+_100871625
|
5.781
|
|
LOC256880
|
uncharacterized LOC256880
|
|
chr19_+_13049413
|
5.754
|
NM_004343
|
CALR
|
calreticulin
|
|
chr17_-_36831156
|
5.739
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr18_-_21017816
|
5.730
|
NM_032933
|
TMEM241
|
transmembrane protein 241
|
|
chr9_-_140009171
|
5.713
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr17_-_74722614
|
5.701
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr7_-_27135524
|
5.696
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_+_102074002
|
5.666
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr19_-_48018163
|
5.652
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr11_-_798196
|
5.602
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr14_-_65569056
|
5.599
|
|
MAX
|
MYC associated factor X
|
|
chr7_+_48128939
|
5.590
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr19_-_10426673
|
5.588
|
NM_001031734
|
FDX1L
|
ferredoxin 1-like
|
|
chr1_+_26496382
|
5.552
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr1_-_111991859
|
5.539
|
|
WDR77
|
WD repeat domain 77
|
|
chr16_+_57023411
|
5.534
|
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr1_+_180123975
|
5.502
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr19_+_49458151
|
5.470
|
|
BAX
|
BCL2-associated X protein
|
|
chr17_+_40719130
|
5.466
|
|
MLX
|
MAX-like protein X
|
|
chr10_-_32217762
|
5.419
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr1_-_226187055
|
5.406
|
NM_152608
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr8_+_67341244
|
5.381
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr1_+_180123906
|
5.374
|
NM_001004128
NM_002826
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr5_-_133706717
|
5.354
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr9_-_140009167
|
5.348
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr3_-_113465101
|
5.344
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chrX_+_54556633
|
5.324
|
NM_001184819
NM_019067
|
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like
|
|
chr14_+_62162042
|
5.316
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr19_-_12833461
|
5.300
|
NM_001136196
|
TNPO2
|
transportin 2
|
|
chr4_-_57301643
|
5.299
|
NM_002703
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr5_+_110427869
|
5.285
|
NM_139281
|
WDR36
|
WD repeat domain 36
|
|
chr6_-_7313352
|
5.282
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr9_-_34048887
|
5.258
|
NM_018449
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr6_+_36562135
|
5.256
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr4_-_2935851
|
5.254
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr6_-_28891409
|
5.248
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr7_-_960509
|
5.246
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr6_+_10695155
|
5.239
|
NM_017906
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr1_+_180124031
|
5.216
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr7_+_4815277
|
5.202
|
|
KIAA0415
|
KIAA0415
|
|
chr5_-_172755195
|
5.201
|
|
STC2
|
stanniocalcin 2
|
|
chr12_-_48152180
|
5.160
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr2_+_26395959
|
5.154
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr10_+_70715883
|
5.137
|
NM_004728
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 21
|
|
chr12_+_54519836
|
5.130
|
|
LOC400043
|
uncharacterized LOC400043
|
|
chr8_+_98656428
|
5.098
|
|
MTDH
|
metadherin
|
|
chr8_-_104427319
|
5.089
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr1_-_11120081
|
5.087
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr19_+_10764987
|
5.073
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr9_-_140009158
|
5.060
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr3_+_50654600
|
5.053
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr1_+_180124023
|
5.053
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr21_+_45209398
|
5.043
|
NM_003683
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr22_+_35777087
|
5.028
|
|
HMOX1
|
heme oxygenase (decycling) 1
|