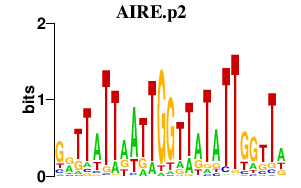
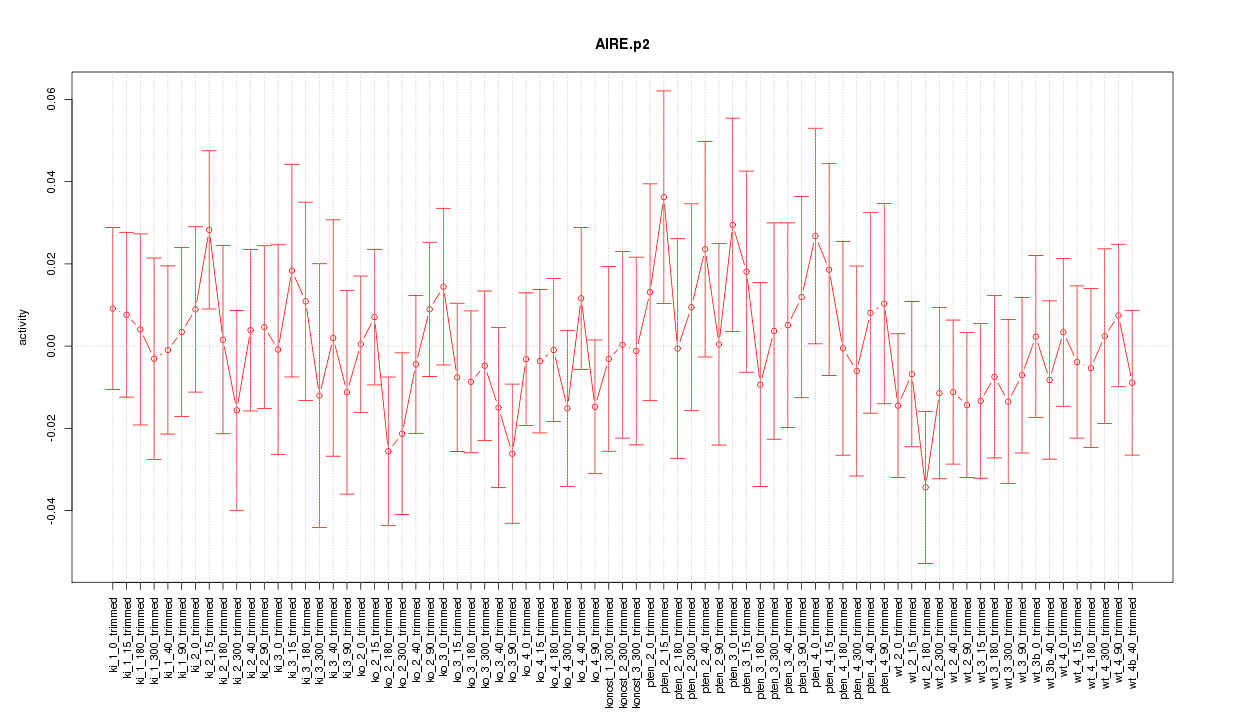
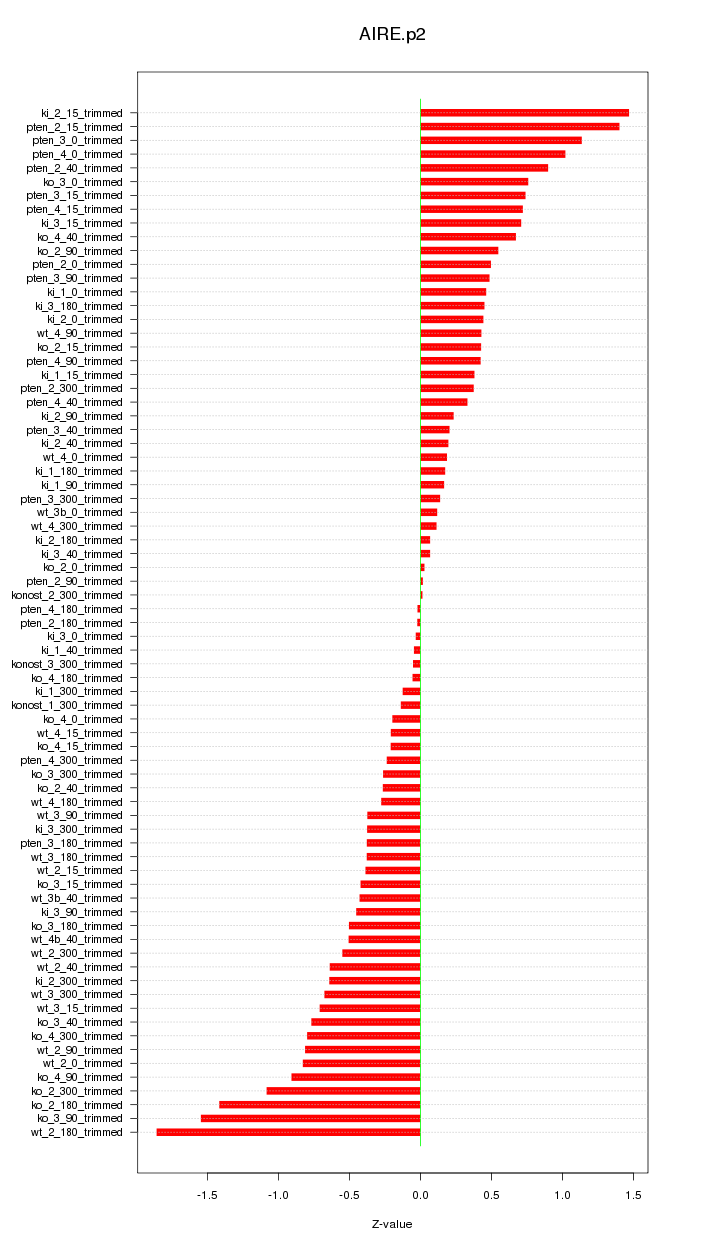
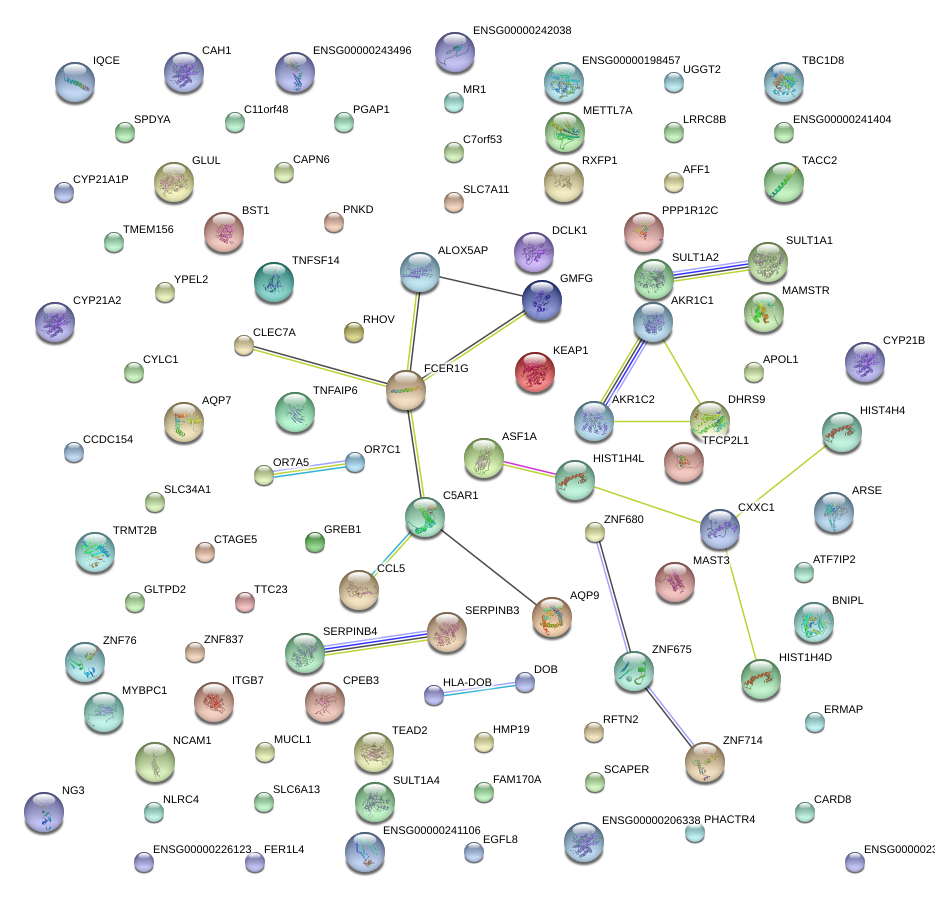
|
chr17_+_4692226
|
3.134
|
NM_001014985
|
GLTPD2
|
glycolipid transfer protein domain containing 2
|
|
chr2_+_29038862
|
3.039
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr10_+_123748688
|
2.922
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr4_+_159442865
|
2.799
|
NM_001253727
NM_001253728
NM_001253729
NM_001253730
NM_001253732
NM_001253733
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr12_-_53600999
|
2.511
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr12_+_101988746
|
2.406
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr15_+_58430394
|
2.178
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr18_-_61329113
|
1.841
|
NM_006919
|
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
|
|
chr19_-_14939275
|
1.803
|
NM_017506
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5
|
|
chr12_-_10282680
|
1.650
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr1_+_161185084
|
1.563
|
NM_004106
|
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide
|
|
chr12_-_371994
|
1.522
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr13_+_31309644
|
1.501
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chrX_-_2882288
|
1.491
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chrX_-_110513665
|
1.487
|
|
CAPN6
|
calpain 6
|
|
chr5_+_176811431
|
1.480
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr1_+_181002560
|
1.456
|
NM_001194999
NM_001195000
NM_001195035
NM_001531
|
MR1
|
major histocompatibility complex, class I-related
|
|
chr19_-_58892382
|
1.427
|
NM_001129730
NM_138466
|
ZNF837
|
zinc finger protein 837
|
|
chr15_-_99789585
|
1.408
|
NM_001040655
NM_001040656
NM_001040657
NM_001040658
NM_001040659
NM_001040660
NM_022905
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr11_+_112832089
|
1.396
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr22_+_36649172
|
1.307
|
|
APOL1
|
apolipoprotein L, 1
|
|
chr19_-_49220213
|
1.291
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr22_+_36649116
|
1.248
|
NM_001136540
NM_001136541
NM_003661
NM_145343
|
APOL1
|
apolipoprotein L, 1
|
|
chr7_+_112120907
|
1.237
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr5_+_118965253
|
1.231
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chr10_-_94003002
|
1.215
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chrX_+_83116153
|
1.205
|
NM_021118
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1
|
|
chr7_+_1609730
|
1.197
|
|
KIAA1908
|
uncharacterized LOC114796
|
|
chr16_+_10522724
|
1.153
|
NM_024997
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr19_-_49865638
|
1.099
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr17_-_34207308
|
1.058
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr12_+_51318743
|
1.055
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr18_-_47814691
|
1.048
|
NM_001101654
NM_014593
|
CXXC1
|
CXXC finger protein 1
|
|
chr19_-_48752921
|
1.033
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr1_+_161185061
|
1.028
|
|
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide
|
|
chr2_-_198540583
|
1.027
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr6_+_32132379
|
1.009
|
NM_030652
|
EGFL8
|
EGF-like-domain, multiple 8
|
|
chr6_+_119215240
|
1.004
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr6_+_35226685
|
1.001
|
|
ZNF76
|
zinc finger protein 76
|
|
chr2_-_122042767
|
0.971
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr4_-_39033981
|
0.962
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr19_-_6670078
|
0.950
|
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr12_-_57693146
|
0.941
|
|
|
|
|
chr7_-_64023454
|
0.926
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr6_-_99849266
|
0.885
|
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr14_+_39734475
|
0.885
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr19_+_21264952
|
0.880
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr13_-_36429798
|
0.867
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr19_-_55628780
|
0.866
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C
|
|
chr13_+_31309690
|
0.863
|
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr7_-_64467006
|
0.860
|
NM_001007253
|
ERV3-1
|
endogenous retrovirus group 3, member 1
|
|
chrM_+_9182
|
0.858
|
|
|
|
|
chr19_-_39826676
|
0.853
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr19_+_47813074
|
0.852
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr6_-_26189303
|
0.852
|
NM_003539
|
HIST1H4D
|
histone cluster 1, H4d
|
|
chr19_+_18208596
|
0.834
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr1_+_43282775
|
0.822
|
NM_001017922
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chrM_+_9217
|
0.822
|
|
|
|
|
chr19_-_48759141
|
0.819
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr2_+_152214105
|
0.809
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr16_-_28621279
|
0.776
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr2_+_11680079
|
0.770
|
NM_148903
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr20_-_34195409
|
0.765
|
|
FER1L4
|
fer-1-like 4 (C. elegans) pseudogene
|
|
chr10_+_5005453
|
0.748
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr2_-_101767714
|
0.745
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr1_+_151009028
|
0.727
|
NM_001159642
NM_138278
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like
|
|
chr16_-_1494489
|
0.726
|
NM_001143980
|
CCDC154
|
coiled-coil domain containing 154
|
|
chrX_-_100307042
|
0.723
|
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chrM_+_8857
|
0.708
|
|
|
|
|
chr2_-_32490688
|
0.707
|
NM_001199139
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr6_+_29759682
|
0.696
|
NM_001207043
|
LOC554223
|
histocompatibility antigen-related
|
|
chr19_-_23869939
|
0.694
|
NM_138330
|
ZNF675
|
zinc finger protein 675
|
|
chrX_-_100307091
|
0.693
|
NM_001167970
NM_001167971
NM_001167972
NM_024917
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr19_-_10613767
|
0.691
|
NM_203500
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr10_-_43187174
|
0.690
|
|
|
|
|
chr15_-_41166336
|
0.678
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr1_+_28764660
|
0.676
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr11_-_62439085
|
0.674
|
|
C11orf48
|
chromosome 11 open reading frame 48
|
|
chr18_-_61311465
|
0.672
|
NM_002974
|
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr4_+_87968191
|
0.663
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr7_+_2598712
|
0.656
|
|
IQCE
|
IQ motif containing E
|
|
chr5_+_173472692
|
0.642
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr12_+_55248298
|
0.640
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chr4_+_15704752
|
0.638
|
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr4_-_139163222
|
0.638
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr6_+_31973358
|
0.637
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr7_+_2598676
|
0.634
|
|
IQCE
|
IQ motif containing E
|
|
chr15_-_77154284
|
0.628
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr1_-_182357871
|
0.626
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr2_+_169929009
|
0.619
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr7_+_1609698
|
0.610
|
|
KIAA1908
|
uncharacterized LOC114796
|
|
chr6_-_32784727
|
0.607
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr16_+_2809933
|
0.602
|
|
|
|
|
chr10_+_135207607
|
0.591
|
NM_138384
|
MTG1
|
mitochondrial GTPase 1 homolog (S. cerevisiae)
|
|
chr14_+_95078713
|
0.590
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr2_-_188312872
|
0.588
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr19_-_39926535
|
0.562
|
NM_001020
|
RPS16
|
ribosomal protein S16
|
|
chr7_-_102283237
|
0.551
|
NM_001114403
|
UPK3BL
|
uroplakin 3B-like
|
|
chr18_-_65182507
|
0.549
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr19_-_46285752
|
0.547
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr3_-_93747453
|
0.547
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chrM_-_9203
|
0.536
|
|
|
|
|
chr5_+_60933635
|
0.534
|
NM_173667
|
C5orf64
|
chromosome 5 open reading frame 64
|
|
chr9_+_42717233
|
0.528
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr7_+_76109826
|
0.520
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr11_+_114549199
|
0.517
|
NM_182495
|
FAM55B
|
family with sequence similarity 55, member B
|
|
chr17_-_48945338
|
0.514
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr10_+_5238756
|
0.514
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr19_+_3728373
|
0.499
|
NM_014428
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chrM_+_4770
|
0.490
|
|
|
|
|
chr6_-_13295817
|
0.489
|
NM_001242698
|
LOC100130357
|
uncharacterized LOC100130357
|
|
chr2_+_12858363
|
0.484
|
|
|
|
|
chr22_-_36556781
|
0.484
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chrX_-_100306859
|
0.482
|
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr18_+_52258389
|
0.479
|
NM_173629
|
C18orf26
|
chromosome 18 open reading frame 26
|
|
chr1_-_161207879
|
0.478
|
NM_001077469
NM_001077470
NM_001077471
NM_001077472
NM_001077473
NM_001077474
NM_001077475
NM_001077476
NM_001077477
NM_001077478
NM_001077479
NM_001077480
NM_001077481
NM_001077482
NM_005122
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr16_+_67001377
|
0.477
|
NM_001185176
|
CES3
|
carboxylesterase 3
|
|
chr15_+_66585632
|
0.476
|
NM_133375
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like
|
|
chr12_-_8043743
|
0.476
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr16_-_4817124
|
0.474
|
NM_021646
|
ZNF500
|
zinc finger protein 500
|
|
chr12_+_8332789
|
0.464
|
|
FAM66C
|
family with sequence similarity 66, member C
|
|
chr6_-_138189313
|
0.459
|
|
|
|
|
chr3_-_157217342
|
0.458
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr6_+_33244916
|
0.456
|
NM_003782
|
B3GALT4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4
|
|
chr22_-_22089975
|
0.455
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr15_+_41576150
|
0.452
|
|
OIP5-AS1
|
OIP5 antisense RNA 1 (non-protein coding)
|
|
chr19_-_10614083
|
0.450
|
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr3_-_194119994
|
0.448
|
NM_004488
|
GP5
|
glycoprotein V (platelet)
|
|
chr16_+_66995099
|
0.447
|
NM_001185177
NM_024922
|
CES3
|
carboxylesterase 3
|
|
chr9_-_70429730
|
0.447
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr20_-_42939737
|
0.446
|
NM_001080472
|
FITM2
|
fat storage-inducing transmembrane protein 2
|
|
chr19_+_48248792
|
0.446
|
NM_015710
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr19_+_48248798
|
0.440
|
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr2_-_170430312
|
0.440
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr3_+_132036210
|
0.431
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr3_+_107096187
|
0.430
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr10_-_61122219
|
0.429
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr19_-_39322411
|
0.425
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr6_-_133078894
|
0.422
|
NM_001242350
NM_004665
|
VNN2
|
vanin 2
|
|
chr16_-_787888
|
0.421
|
|
NARFL
|
nuclear prelamin A recognition factor-like
|
|
chr2_-_214015053
|
0.414
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr16_-_86588680
|
0.413
|
NM_001159380
NM_001159377
NM_001159378
NM_001159379
NM_022764
|
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing
|
|
chr9_-_70178814
|
0.408
|
NM_001126334
|
FOXD4L5
|
forkhead box D4-like 5
|
|
chr7_-_97601654
|
0.408
|
|
|
|
|
chr19_+_7599037
|
0.406
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr12_+_51318515
|
0.404
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr16_-_4524895
|
0.403
|
NM_020677
|
NMRAL1
|
NmrA-like family domain containing 1
|
|
chr2_+_234590583
|
0.395
|
NM_019077
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7
|
|
chr5_+_173472737
|
0.388
|
|
HMP19
|
HMP19 protein
|
|
chr5_-_177207465
|
0.387
|
NM_173663
|
FAM153A
|
family with sequence similarity 153, member A
|
|
chr2_-_61245363
|
0.387
|
NM_144709
|
PUS10
|
pseudouridylate synthase 10
|
|
chr9_-_136933552
|
0.381
|
|
BRD3
|
bromodomain containing 3
|
|
chr6_-_53213747
|
0.377
|
NM_001242828
NM_001242830
NM_001242831
NM_021814
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chrM_+_9725
|
0.376
|
|
|
|
|
chr2_-_190927321
|
0.373
|
NM_005259
|
MSTN
|
myostatin
|
|
chr7_+_2598646
|
0.371
|
|
IQCE
|
IQ motif containing E
|
|
chr4_-_159093523
|
0.370
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr12_-_123187842
|
0.367
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr9_-_95298251
|
0.366
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chrM_+_4465
|
0.362
|
|
|
|
|
chr11_+_65479685
|
0.351
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr19_-_8408120
|
0.349
|
NM_198471
|
KANK3
|
KN motif and ankyrin repeat domains 3
|
|
chr17_-_67138013
|
0.347
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr11_-_64578765
|
0.344
|
NM_130803
NM_130804
|
MEN1
|
multiple endocrine neoplasia I
|
|
chr1_-_169680677
|
0.339
|
NM_000655
|
SELL
|
selectin L
|
|
chr12_-_56753856
|
0.338
|
|
STAT2
|
signal transducer and activator of transcription 2, 113kDa
|
|
chr3_+_189507448
|
0.335
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr3_+_29322802
|
0.333
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr6_+_31590468
|
0.333
|
|
|
|
|
chr15_+_66585822
|
0.330
|
NM_001143688
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like
|
|
chr7_+_76090971
|
0.329
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr1_+_210501595
|
0.327
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr17_+_41003184
|
0.326
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr12_-_56753912
|
0.322
|
|
STAT2
|
signal transducer and activator of transcription 2, 113kDa
|
|
chr3_-_186262114
|
0.322
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chr6_-_42858526
|
0.318
|
NM_001008739
|
C6orf226
|
chromosome 6 open reading frame 226
|
|
chr6_+_31683084
|
0.318
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr6_-_31830593
|
0.311
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr1_-_216978708
|
0.310
|
NM_001243514
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr22_+_32753853
|
0.309
|
NM_001098535
|
RFPL3
|
ret finger protein-like 3
|
|
chr7_+_74508346
|
0.308
|
NM_001003795
|
GTF2IRD2B
GTF2IRD2
|
GTF2I repeat domain containing 2B
GTF2I repeat domain containing 2
|
|
chr12_-_53558276
|
0.304
|
NM_001244706
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr4_+_39408472
|
0.296
|
NM_175737
|
KLB
|
klotho beta
|
|
chr2_-_190627923
|
0.294
|
NM_022353
|
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1
|
|
chr3_+_46283871
|
0.288
|
NM_001164680
NM_001837
NM_178328
NM_178329
|
CCR3
|
chemokine (C-C motif) receptor 3
|
|
chr11_+_5641173
|
0.286
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr5_+_43609261
|
0.285
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr7_+_2598611
|
0.283
|
NM_001100390
NM_152558
|
IQCE
|
IQ motif containing E
|
|
chr19_-_46526555
|
0.282
|
NM_005091
|
PGLYRP1
|
peptidoglycan recognition protein 1
|
|
chr4_+_170581212
|
0.282
|
NM_001243374
|
CLCN3
|
chloride channel 3
|
|
chr22_-_17302555
|
0.282
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr11_-_6462129
|
0.278
|
NM_000613
|
HPX
|
hemopexin
|
|
chr21_-_35884503
|
0.274
|
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr12_-_54778456
|
0.272
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr12_+_62954275
|
0.270
|
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr7_-_74267824
|
0.267
|
NM_173537
|
GTF2IRD2B
GTF2IRD2
|
GTF2I repeat domain containing 2B
GTF2I repeat domain containing 2
|
|
chr6_+_26365397
|
0.267
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr17_+_65936590
|
0.264
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr16_-_3627353
|
0.263
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr15_-_55800431
|
0.262
|
NM_001033559
NM_001033560
NM_130810
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|