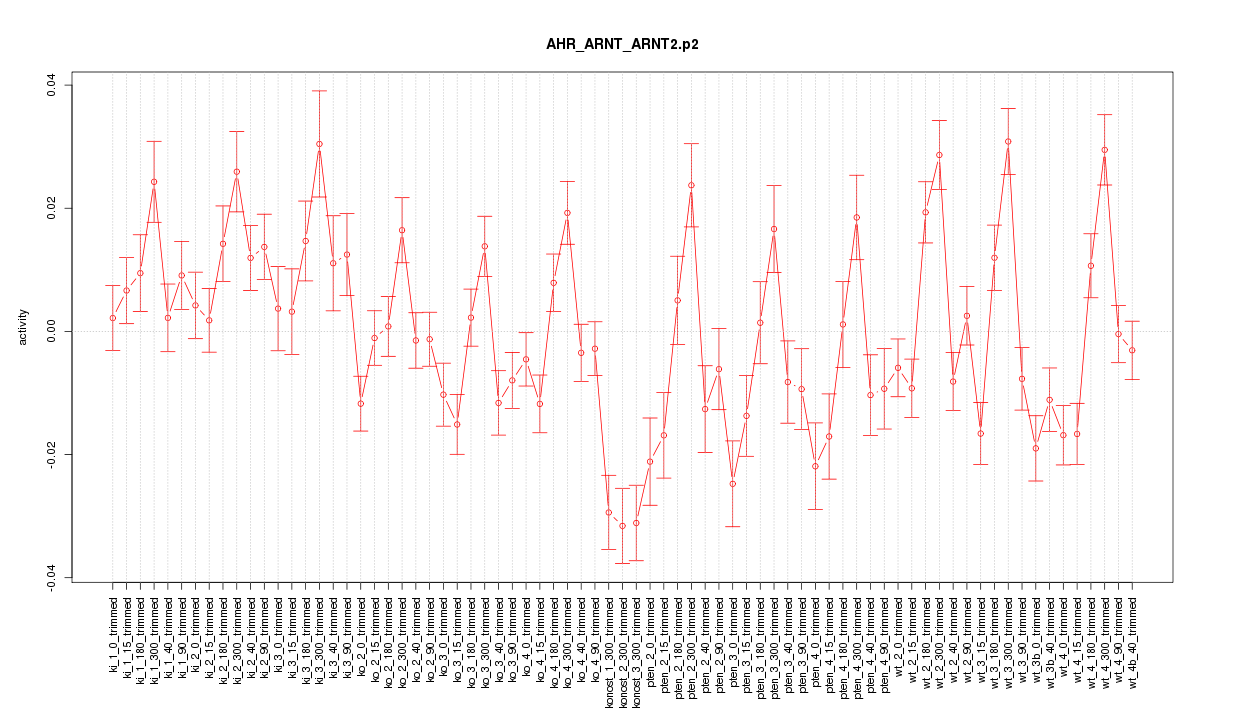
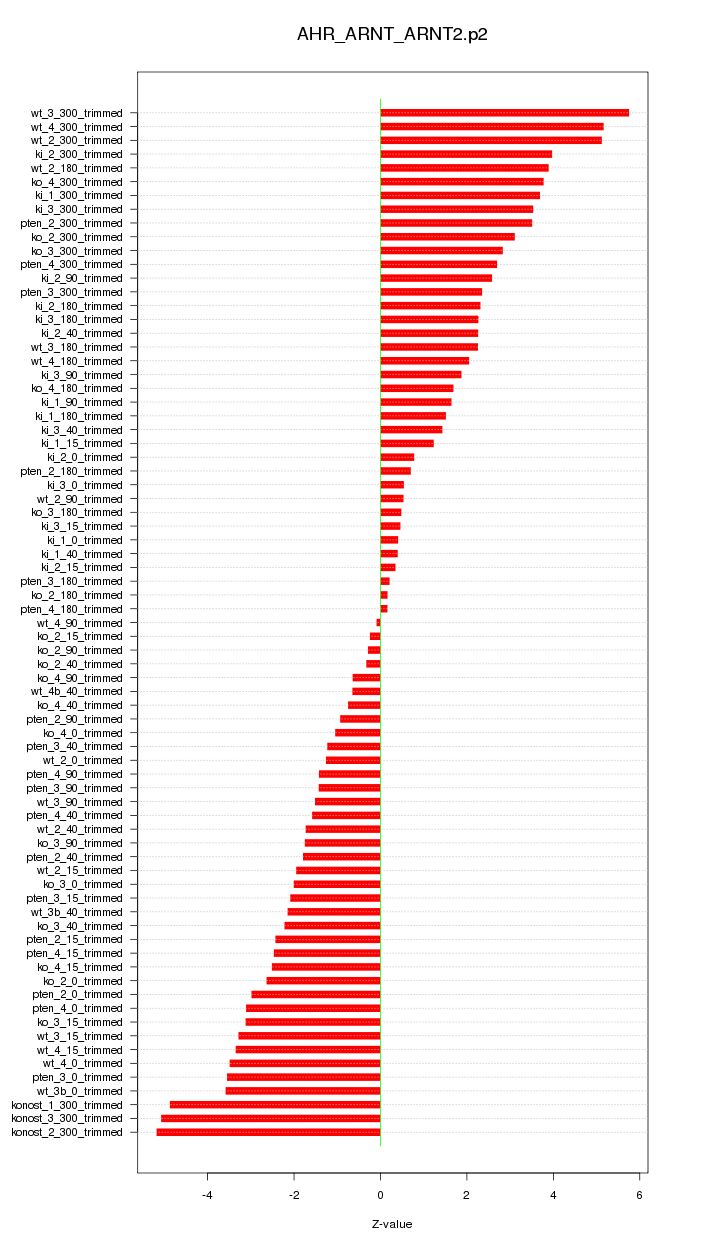
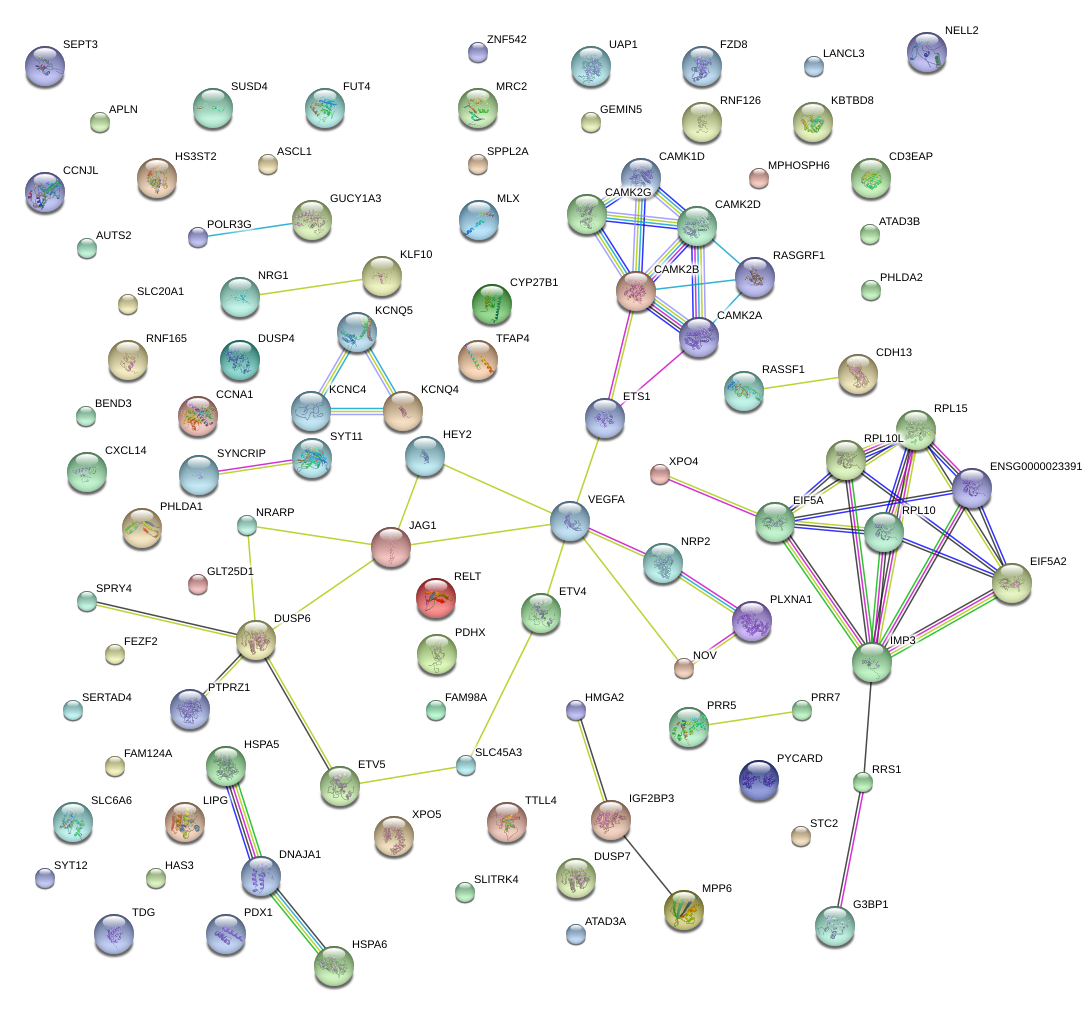
|
chr12_-_89745753
|
14.222
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr15_-_75932509
|
13.464
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr22_+_42372763
|
13.224
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr11_+_66790815
|
12.331
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr1_+_1407112
|
12.261
|
|
ATAD3B
|
ATPase family, AAA domain containing 3B
|
|
chr1_-_205649616
|
12.105
|
NM_033102
|
SLC45A3
|
solute carrier family 45, member 3
|
|
chr1_+_1407158
|
11.813
|
NM_031921
|
ATAD3B
|
ATPase family, AAA domain containing 3B
|
|
chr13_+_28494136
|
11.143
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr1_+_210406194
|
11.113
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr16_+_69139466
|
11.048
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr4_+_156588811
|
10.411
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr12_-_45270071
|
10.275
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_-_205649537
|
10.084
|
|
SLC45A3
|
solute carrier family 45, member 3
|
|
chr12_-_76424474
|
9.934
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr19_+_45909466
|
9.504
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr6_+_43738757
|
9.456
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_-_223537400
|
8.691
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr6_+_73331808
|
8.637
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr5_-_141704575
|
8.598
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr1_+_162531349
|
8.557
|
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chrX_+_37431015
|
8.421
|
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|
|
chr5_-_141704468
|
8.411
|
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr17_-_41623246
|
8.358
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr12_-_45270157
|
8.262
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_50374875
|
8.254
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr5_+_151151462
|
8.245
|
NM_005754
NM_198395
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr1_+_1447537
|
8.194
|
|
ATAD3A
ATAD3B
|
ATPase family, AAA domain containing 3A
ATPase family, AAA domain containing 3B
|
|
chr5_+_151151514
|
8.159
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr6_+_126070813
|
8.103
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr15_-_79383080
|
8.021
|
NM_001145648
NM_002891
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr1_+_1447495
|
8.014
|
NM_001170535
NM_018188
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr3_+_14444062
|
7.867
|
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr3_-_185826748
|
7.816
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr1_+_161494035
|
7.704
|
NM_002155
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B')
|
|
chr6_-_107435635
|
7.646
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr3_-_62359189
|
7.595
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr19_+_17666502
|
7.568
|
NM_024656
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr5_+_151151516
|
7.500
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr1_+_1407157
|
7.455
|
|
|
|
|
chr3_-_50374795
|
7.387
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr18_+_47088400
|
7.387
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_+_1447871
|
7.372
|
NM_001170536
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr5_-_172755195
|
7.297
|
|
STC2
|
stanniocalcin 2
|
|
chr16_+_22825806
|
7.221
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr6_-_43543600
|
7.193
|
|
XPO5
|
exportin 5
|
|
chr6_-_43543749
|
7.187
|
|
XPO5
|
exportin 5
|
|
chr2_+_113403433
|
7.165
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr17_-_41623652
|
7.077
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr2_-_33824294
|
6.976
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr6_-_43543767
|
6.968
|
NM_020750
|
XPO5
|
exportin 5
|
|
chr19_+_17666381
|
6.961
|
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr9_-_140196702
|
6.882
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr16_+_82660573
|
6.867
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr7_+_70231163
|
6.853
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr5_+_151151483
|
6.812
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr11_-_128392061
|
6.809
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr19_+_17666457
|
6.778
|
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr5_+_151151511
|
6.725
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr5_-_172755418
|
6.654
|
|
STC2
|
stanniocalcin 2
|
|
chr16_+_82660641
|
6.641
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr7_+_24612714
|
6.620
|
NM_016447
|
MPP6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6)
|
|
chr5_-_134914447
|
6.493
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr8_+_31497267
|
6.430
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chrX_-_142722869
|
6.402
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr11_-_2950593
|
6.277
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr10_-_35930357
|
6.254
|
NM_031866
|
FZD8
|
frizzled family receptor 8
|
|
chr5_+_176873795
|
6.212
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr19_-_663219
|
6.199
|
NM_194460
|
RNF126
|
ring finger protein 126
|
|
chr8_+_120428551
|
6.193
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr3_+_67048726
|
6.177
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr12_-_45269953
|
6.114
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_45267068
|
6.094
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr2_+_219575507
|
6.090
|
NM_014640
|
TTLL4
|
tubulin tyrosine ligase-like family, member 4
|
|
chr11_+_94277566
|
6.085
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr12_+_66218879
|
6.084
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr5_-_159739482
|
6.055
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr6_+_73331570
|
6.045
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr19_-_663158
|
5.993
|
|
RNF126
|
ring finger protein 126
|
|
chr12_+_104359557
|
5.958
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr1_+_162531229
|
5.909
|
NM_003115
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chr5_-_154317653
|
5.896
|
NM_001252156
NM_015465
|
GEMIN5
|
gem (nuclear organelle) associated protein 5
|
|
chr20_-_1165116
|
5.874
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr6_+_73331992
|
5.866
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chrX_-_128788913
|
5.861
|
NM_017413
|
APLN
|
apelin
|
|
chr19_-_663155
|
5.796
|
|
RNF126
|
ring finger protein 126
|
|
chr2_+_219575684
|
5.782
|
|
TTLL4
|
tubulin tyrosine ligase-like family, member 4
|
|
chr19_-_663195
|
5.724
|
|
RNF126
|
ring finger protein 126
|
|
chr1_+_110753910
|
5.704
|
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr12_-_76425207
|
5.701
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_+_12391728
|
5.691
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr3_-_50374749
|
5.685
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr18_+_43913906
|
5.676
|
|
RNF165
|
ring finger protein 165
|
|
chr8_-_29206321
|
5.597
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr20_-_10654429
|
5.494
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr13_+_37006408
|
5.479
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr3_-_170626425
|
5.452
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr19_-_7968426
|
5.449
|
|
|
|
|
chr11_+_73087670
|
5.415
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr17_+_40719130
|
5.409
|
|
MLX
|
MAX-like protein X
|
|
chr12_+_66218568
|
5.378
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr7_-_44365019
|
5.304
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr13_+_51796363
|
5.290
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr3_-_52090090
|
5.276
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr7_+_121513158
|
5.259
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr12_-_58159386
|
5.254
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr12_-_45269340
|
5.209
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr15_-_79382839
|
5.085
|
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr10_+_12391708
|
5.081
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr17_-_41622727
|
5.041
|
|
ETV4
|
ets variant 4
|
|
chr19_+_56879687
|
5.000
|
|
ZNF542
|
zinc finger protein 542
|
|
chr2_+_206547131
|
4.999
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr16_-_31214054
|
4.985
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chr17_+_60705042
|
4.964
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr16_+_22825481
|
4.963
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr9_-_128003615
|
4.963
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr3_-_170626381
|
4.960
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr8_-_103667882
|
4.957
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr22_+_45098046
|
4.944
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr6_+_126070724
|
4.907
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr16_-_4322695
|
4.889
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr3_+_14444083
|
4.854
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr6_-_86352982
|
4.845
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr3_-_185826332
|
4.826
|
|
ETV5
|
ets variant 5
|
|
chr13_-_21476895
|
4.822
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr16_-_31214003
|
4.821
|
|
PYCARD
|
PYD and CARD domain containing
|
|
chr5_+_89770680
|
4.814
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr8_+_67341244
|
4.811
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr1_+_2518464
|
4.810
|
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr12_+_103351451
|
4.784
|
NM_004316
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr9_+_33025302
|
4.750
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr2_+_220283098
|
4.740
|
NM_001927
|
DES
|
desmin
|
|
chr12_+_4382882
|
4.736
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr13_+_37005966
|
4.734
|
NM_001111046
NM_001111047
|
CCNA1
|
cyclin A1
|
|
chr7_-_150924220
|
4.702
|
NM_005692
NM_007189
|
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr11_-_61348290
|
4.689
|
NM_001252065
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr6_+_126070738
|
4.686
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr12_-_28122884
|
4.683
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr9_-_128003348
|
4.667
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr10_+_105156430
|
4.654
|
|
PDCD11
|
programmed cell death 11
|
|
chr17_-_16472463
|
4.630
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr6_-_86352635
|
4.617
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_110753313
|
4.598
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr19_+_4909447
|
4.579
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr9_+_33025270
|
4.545
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr17_+_54671059
|
4.522
|
NM_005450
|
NOG
|
noggin
|
|
chr15_-_63673918
|
4.504
|
|
CA12
|
carbonic anhydrase XII
|
|
chr10_+_105156415
|
4.490
|
|
PDCD11
|
programmed cell death 11
|
|
chr12_-_49365545
|
4.433
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr3_-_45267620
|
4.432
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr17_-_38256548
|
4.424
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr11_+_129245834
|
4.406
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr16_-_4323000
|
4.395
|
NM_003223
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr17_-_56084514
|
4.389
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr1_+_27719151
|
4.378
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr1_+_221052845
|
4.353
|
|
HLX
|
H2.0-like homeobox
|
|
chr15_-_63674074
|
4.349
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chrX_-_17879355
|
4.341
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr1_+_92495484
|
4.324
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr1_+_9648931
|
4.324
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr10_-_92617670
|
4.323
|
NM_000872
NM_019859
NM_019860
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr10_+_105156382
|
4.293
|
NM_014976
|
PDCD11
|
programmed cell death 11
|
|
chr14_+_103800678
|
4.247
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr1_+_2518071
|
4.230
|
NM_001195736
NM_001195737
NM_001195738
NM_001195740
NM_001195741
NM_152371
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr3_-_127541975
|
4.226
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr20_-_22564893
|
4.225
|
|
FOXA2
|
forkhead box A2
|
|
chr16_-_4323055
|
4.196
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr5_-_134914968
|
4.175
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr20_-_31172874
|
4.171
|
|
LOC284804
|
uncharacterized LOC284804
|
|
chr1_+_221053000
|
4.164
|
|
HLX
|
H2.0-like homeobox
|
|
chr16_+_3019341
|
4.147
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr20_+_48599511
|
4.139
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr13_+_37006628
|
4.133
|
|
CCNA1
|
cyclin A1
|
|
chr18_+_55019720
|
4.110
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr19_-_40732563
|
4.104
|
NM_024877
|
CNTD2
|
cyclin N-terminal domain containing 2
|
|
chr7_-_22539795
|
4.084
|
NM_001164460
NM_207342
|
STEAP1B
|
STEAP family member 1B
|
|
chr18_+_77155966
|
4.080
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr5_-_6633347
|
4.072
|
NM_001193455
NM_017755
|
NSUN2
|
NOP2/Sun domain family, member 2
|
|
chr15_+_38544119
|
4.064
|
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr17_-_38256905
|
4.052
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr1_+_41249479
|
4.048
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr3_-_184870727
|
4.044
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chrX_+_153991106
|
4.028
|
|
DKC1
|
dyskeratosis congenita 1, dyskerin
|
|
chr6_-_86352656
|
4.024
|
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr9_+_99212379
|
4.019
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr7_+_121513210
|
4.012
|
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr12_+_110152186
|
4.010
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr19_-_36980336
|
4.001
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chrX_-_63425484
|
3.968
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr6_-_86352650
|
3.964
|
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr19_-_10341947
|
3.963
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr16_-_15188089
|
3.944
|
NM_018427
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae)
|
|
chr17_-_56084498
|
3.944
|
|
|
|
|
chr17_+_40719064
|
3.943
|
NM_170607
NM_198204
NM_198205
|
MLX
|
MAX-like protein X
|
|
chr6_-_86352564
|
3.939
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr18_+_23806385
|
3.931
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr2_-_160761192
|
3.924
|
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr14_-_69445989
|
3.908
|
|
ACTN1
|
actinin, alpha 1
|
|
chr14_-_69446038
|
3.887
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr17_+_65373362
|
3.886
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chrX_+_37430821
|
3.884
|
NM_001170331
NM_198511
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|