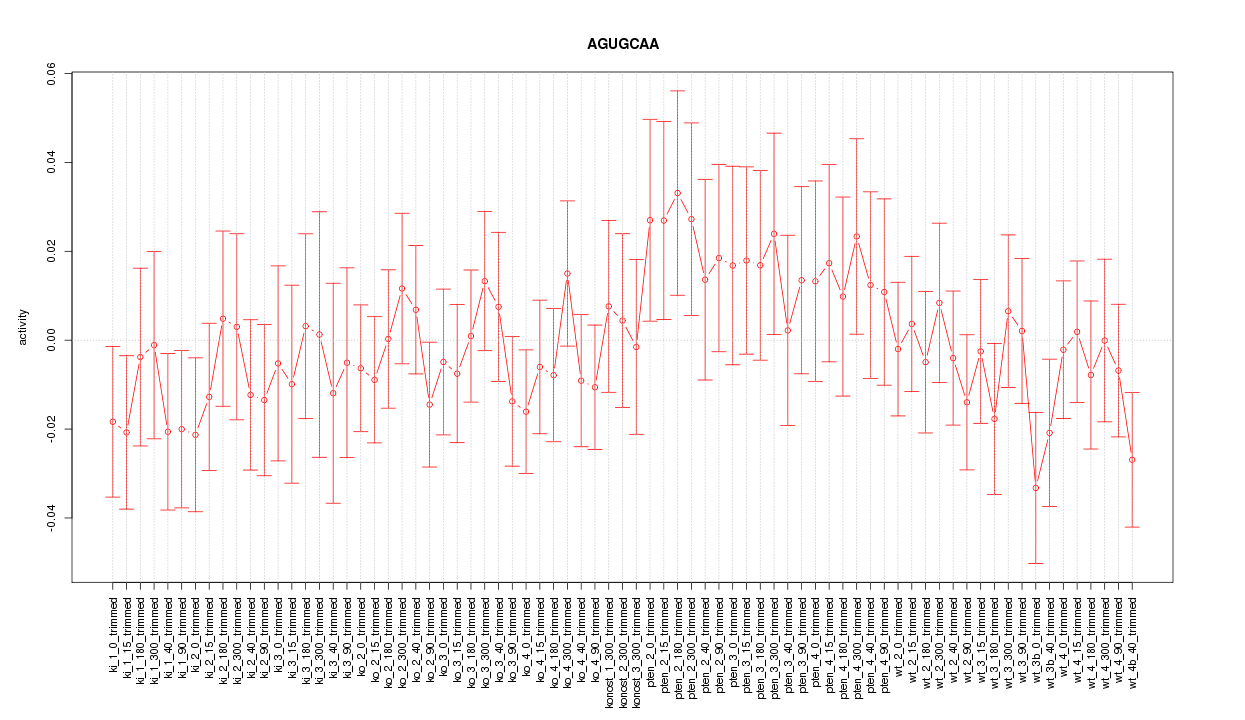
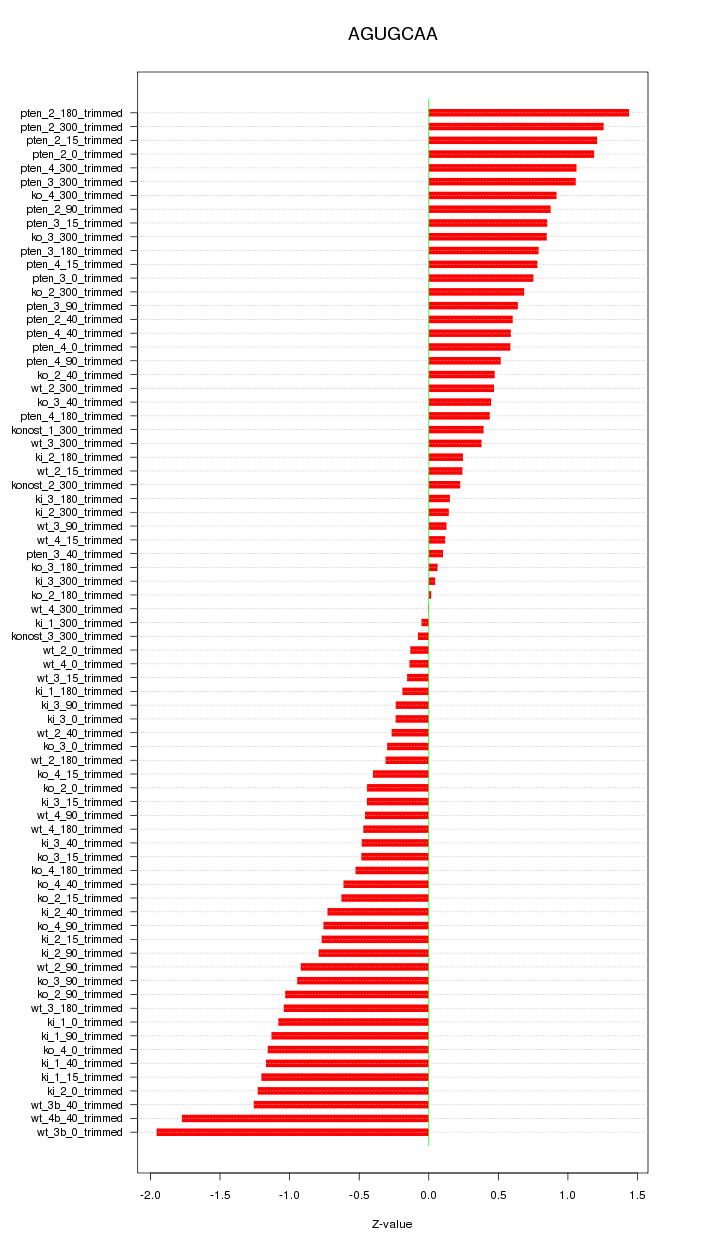
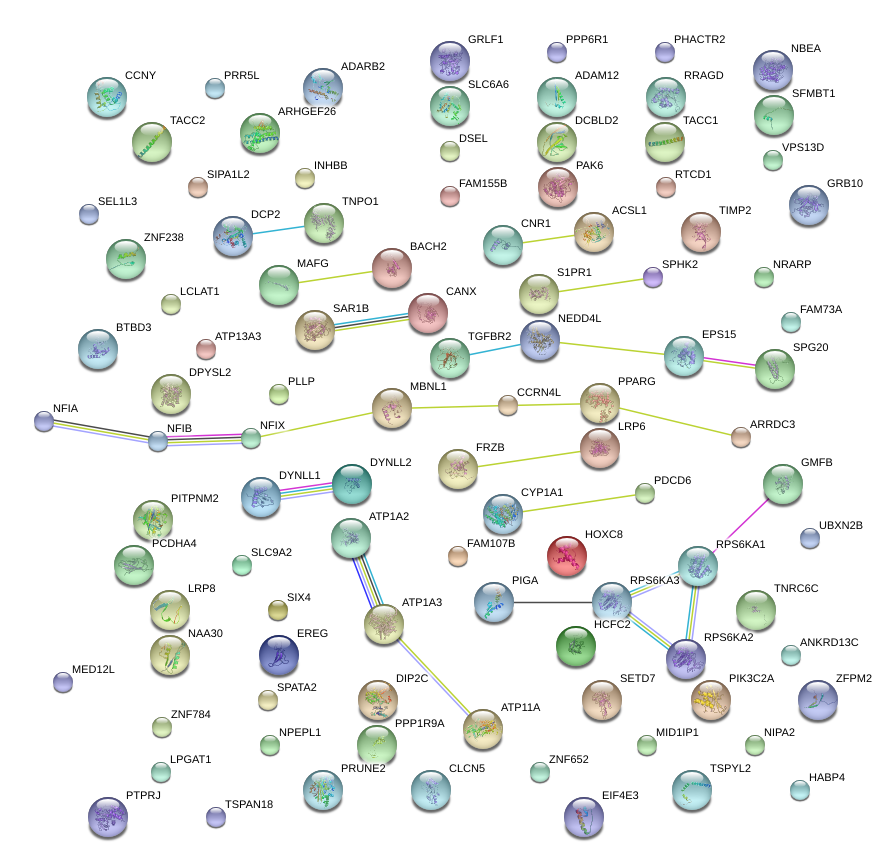
|
chr11_+_44785975
|
8.058
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr4_+_75230856
|
7.982
|
NM_001432
|
EREG
|
epiregulin
|
|
chr10_-_1779669
|
5.464
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr11_+_36317724
|
5.026
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr11_+_36397534
|
4.945
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr11_+_36422546
|
4.443
|
NM_001160169
|
PRR5L
|
proline rich 5 like
|
|
chr18_-_65183922
|
3.051
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr10_+_123923104
|
2.860
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr1_+_101702304
|
2.818
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr2_+_103236147
|
2.565
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr10_+_123748688
|
2.507
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr6_-_167275657
|
2.473
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr10_-_735522
|
2.384
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr1_-_53793743
|
2.363
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr1_-_232651227
|
2.310
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr9_-_79520878
|
2.300
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr6_-_90121966
|
2.224
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr6_-_167040700
|
2.106
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chrX_+_53111482
|
2.067
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr3_-_192635933
|
2.045
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr12_+_54402795
|
1.979
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr2_+_121103670
|
1.929
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr4_-_25864430
|
1.927
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr3_+_14444083
|
1.865
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr20_+_57267818
|
1.844
|
NM_024663
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr19_-_55769920
|
1.830
|
NM_014931
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr20_+_57264186
|
1.822
|
NM_001204872
NM_001204873
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr4_-_185747183
|
1.814
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr2_-_183731279
|
1.772
|
NM_001463
|
FRZB
|
frizzled-related protein
|
|
chrX_+_68725077
|
1.737
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr7_+_94539209
|
1.729
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr7_+_94536926
|
1.695
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chrX_+_49832214
|
1.676
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr1_+_61547388
|
1.642
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr17_+_76000248
|
1.569
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr9_+_99212379
|
1.567
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr16_-_57318574
|
1.553
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chrX_+_38663072
|
1.502
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr13_+_113344354
|
1.489
|
NM_015205
NM_032189
|
ATP11A
|
ATPase, class VI, type 11A
|
|
chr17_-_76921453
|
1.435
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_+_49128208
|
1.419
|
NM_001204160
|
SPHK2
|
sphingosine kinase 2
|
|
chrX_+_38660653
|
1.397
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr18_+_55816546
|
1.396
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_-_50861114
|
1.396
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr10_-_128077072
|
1.384
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr7_-_50800018
|
1.374
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr19_+_47421932
|
1.329
|
NM_004491
|
ARHGAP35
|
Rho GTPase activating protein 35
|
|
chr7_-_50772997
|
1.311
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr18_+_55714457
|
1.281
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_12419702
|
1.266
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr10_-_14816895
|
1.245
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr14_-_61190458
|
1.240
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr18_+_55711388
|
1.238
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr5_+_304290
|
1.221
|
NM_001242412
NM_020731
|
AHRR
|
aryl-hydrocarbon receptor repressor
|
|
chr6_+_143929316
|
1.215
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_+_160085519
|
1.212
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr1_-_51887740
|
1.202
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr3_+_30647901
|
1.198
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr11_-_17191287
|
1.190
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chrX_-_15353675
|
1.190
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr1_+_61542928
|
1.190
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr4_+_166248816
|
1.189
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr5_-_90679115
|
1.183
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr18_+_55862577
|
1.158
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55888750
|
1.157
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_+_150804675
|
1.157
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr1_+_61547625
|
1.151
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_140477290
|
1.146
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chrX_+_49687212
|
1.124
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr15_-_23034406
|
1.121
|
NM_001008860
NM_001008892
NM_001008894
NM_001184888
NM_001184889
NM_030922
|
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2
|
|
chr12_+_104458235
|
1.120
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr14_-_54955714
|
1.112
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr3_-_53079285
|
1.107
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr1_+_244216506
|
1.100
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr9_-_140196702
|
1.098
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr3_-_194188967
|
1.081
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr15_+_40532034
|
1.079
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr3_+_12392993
|
1.075
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr10_+_35535952
|
1.054
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr1_-_70820363
|
1.053
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chr8_+_106331146
|
1.052
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr10_+_35625608
|
1.047
|
NM_145012
|
CCNY
|
cyclin Y
|
|
chr13_+_36050885
|
1.038
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr17_-_47439326
|
1.038
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr5_+_179125796
|
1.030
|
NM_001024649
NM_001746
|
CANX
|
calnexin
|
|
chr11_+_48002063
|
1.022
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr5_-_133968463
|
1.010
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr1_-_212004113
|
1.006
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr1_+_244214552
|
1.005
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_78245306
|
1.004
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr1_+_100731696
|
1.001
|
NM_001130841
NM_003729
|
RTCD1
|
RNA terminal phosphate cyclase domain 1
|
|
chr19_-_56135934
|
0.977
|
NM_203374
|
ZNF784
|
zinc finger protein 784
|
|
chr1_-_51984994
|
0.969
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr7_+_100081549
|
0.967
|
NM_173564
|
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1
|
|
chr17_+_56160726
|
0.961
|
NM_080677
|
DYNLL2
|
dynein, light chain, LC8-type 2
|
|
chr3_-_98620452
|
0.954
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr5_+_112312392
|
0.953
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr22_-_32146119
|
0.952
|
NM_173566
|
PRR14L
|
proline rich 14-like
|
|
chr17_-_79881304
|
0.949
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr3_+_153839136
|
0.935
|
NM_001251963
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr20_-_48530275
|
0.930
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr8_+_38585703
|
0.927
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr6_-_88854989
|
0.925
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr3_+_152017193
|
0.920
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr20_+_11898536
|
0.917
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr8_+_59323822
|
0.916
|
NM_001077619
|
UBXN2B
|
UBX domain protein 2B
|
|
chr14_+_57857261
|
0.916
|
NM_001011713
|
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit
|
|
chr6_-_91006460
|
0.906
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr13_+_35516390
|
0.906
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr2_+_64681270
|
0.898
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr15_+_40509628
|
0.898
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr5_+_72143929
|
0.891
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr2_+_30670082
|
0.890
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr3_+_12329332
|
0.887
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr3_-_71778268
|
0.882
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr13_-_36944289
|
0.882
|
NM_001142294
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr12_-_123594974
|
0.879
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr4_+_139936938
|
0.879
|
NM_012118
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr8_+_26435339
|
0.876
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr1_+_12290056
|
0.875
|
NM_015378
NM_018156
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae)
|
|
chr1_+_113010039
|
0.874
|
NM_004185
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr6_+_44238479
|
0.874
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr4_+_129730772
|
0.869
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr3_-_15839674
|
0.867
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_+_109964086
|
0.855
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr13_-_33780142
|
0.853
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_-_128045995
|
0.850
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr2_+_192542860
|
0.849
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr10_-_27529263
|
0.847
|
NM_145698
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chrX_-_108976509
|
0.844
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr17_+_54911452
|
0.839
|
NM_003647
|
DGKE
|
diacylglycerol kinase, epsilon 64kDa
|
|
chr14_-_95599794
|
0.838
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr1_+_151584514
|
0.831
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr18_-_30050394
|
0.826
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr8_+_38644721
|
0.826
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr3_-_53080038
|
0.824
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr3_-_87040246
|
0.824
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr6_+_143999058
|
0.821
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr5_-_89825292
|
0.820
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr17_+_36861857
|
0.816
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr16_+_19125243
|
0.811
|
NM_001034841
|
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2
|
|
chr12_-_30907447
|
0.809
|
NM_001002259
NM_001206856
NM_023925
NM_032156
|
CAPRIN2
|
caprin family member 2
|
|
chr5_+_96294154
|
0.809
|
NM_175920
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr8_-_12612968
|
0.803
|
NM_152271
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chr1_+_47799447
|
0.801
|
NM_001136140
NM_016308
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chr3_-_113465101
|
0.795
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr8_-_67579446
|
0.786
|
NM_025054
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr3_+_12330336
|
0.785
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr1_-_226926875
|
0.775
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr2_-_172750724
|
0.774
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr11_-_122932844
|
0.761
|
NM_006597
NM_153201
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr6_-_42016609
|
0.760
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr5_+_96271345
|
0.753
|
NM_005575
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr2_-_65357249
|
0.752
|
NM_004161
NM_015543
|
RAB1A
|
RAB1A, member RAS oncogene family
|
|
chr1_+_162466963
|
0.751
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr10_-_81205091
|
0.751
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr1_+_218518675
|
0.744
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr14_-_95608054
|
0.741
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr12_+_121124922
|
0.741
|
NM_014730
|
MLEC
|
malectin
|
|
chrX_+_68835910
|
0.740
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr19_+_49122547
|
0.736
|
NM_001204158
NM_001243876
NM_020126
|
SPHK2
|
sphingosine kinase 2
|
|
chr1_+_57110745
|
0.731
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chrX_-_131623994
|
0.730
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr4_+_38665587
|
0.727
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr3_+_57261729
|
0.721
|
NM_012096
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr3_+_61547179
|
0.719
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chrX_-_103087106
|
0.717
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr17_+_57697036
|
0.708
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chrX_-_135056132
|
0.707
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr7_+_116312412
|
0.705
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr13_-_33760215
|
0.702
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_+_95679075
|
0.699
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr5_-_59783885
|
0.695
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_-_25034064
|
0.693
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr9_-_138987096
|
0.692
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr4_-_53522756
|
0.691
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr2_+_74273449
|
0.687
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chrX_-_131547536
|
0.679
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chrX_+_128674181
|
0.678
|
NM_000276
NM_001587
|
OCRL
|
oculocerebrorenal syndrome of Lowe
|
|
chr17_+_52978022
|
0.678
|
NM_005486
|
TOM1L1
|
target of myb1 (chicken)-like 1
|
|
chr6_-_91006580
|
0.677
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_+_11538486
|
0.672
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr3_-_15798057
|
0.671
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr5_-_65017874
|
0.661
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr12_-_77459198
|
0.658
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr13_-_21476895
|
0.657
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr7_-_27183225
|
0.654
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr1_+_204485461
|
0.650
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr20_-_39317875
|
0.649
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr5_-_141704575
|
0.645
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr12_-_51591949
|
0.639
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr7_-_27166638
|
0.639
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr6_+_64281910
|
0.638
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_+_139259349
|
0.638
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr10_+_98592658
|
0.636
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_+_71403040
|
0.636
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_-_71803513
|
0.636
|
NM_001134649
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr3_-_178789597
|
0.634
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr14_-_95623665
|
0.630
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr10_+_126490353
|
0.630
|
NM_032182
|
FAM175B
|
family with sequence similarity 175, member B
|