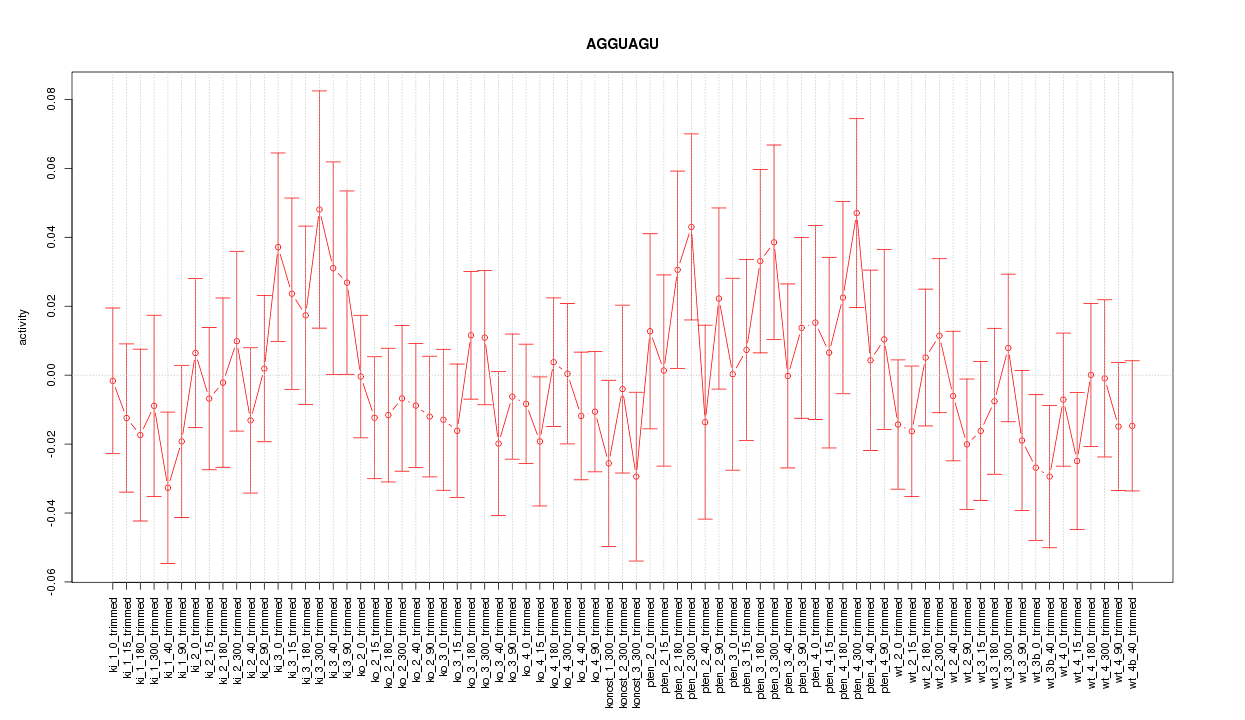
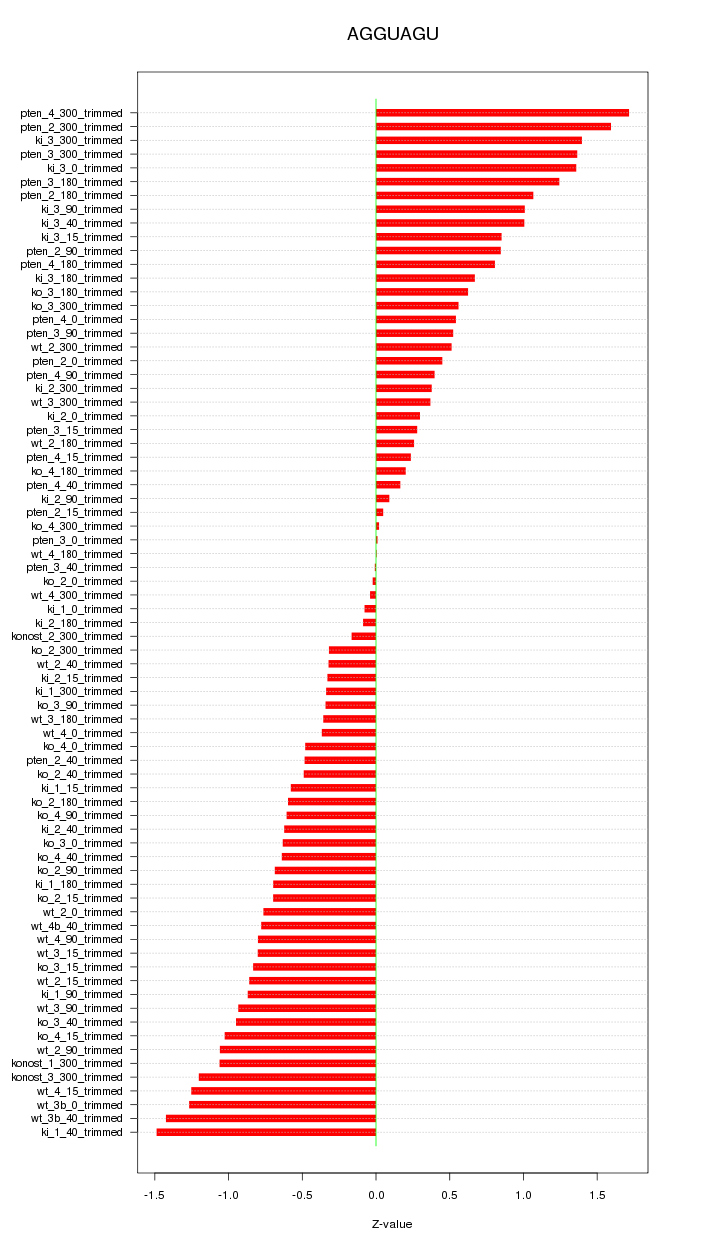
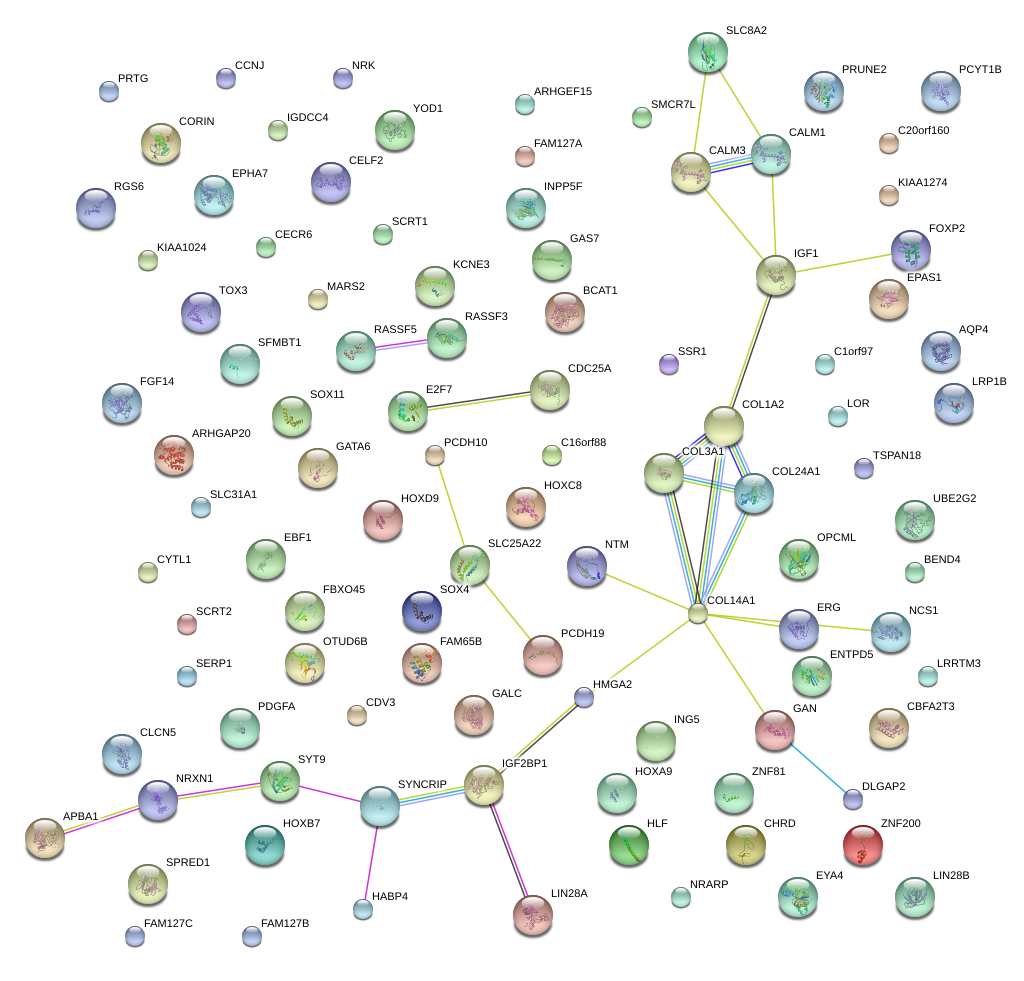
|
chr11_+_44785975
|
9.425
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr12_+_54402795
|
3.191
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr15_-_56035176
|
2.798
|
NM_173814
|
PRTG
|
protogenin
|
|
chr10_+_97803064
|
2.348
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr2_+_198570027
|
2.296
|
NM_138395
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial
|
|
chr17_+_47074749
|
2.214
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr11_-_796175
|
2.207
|
NM_024698
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr6_+_105404922
|
2.201
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr3_-_48229800
|
2.158
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr6_-_24911194
|
2.123
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr4_-_42154894
|
2.117
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr15_-_65715385
|
2.047
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr9_+_99212379
|
2.032
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr14_+_72399149
|
2.000
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr17_-_10101854
|
1.964
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr1_+_153232178
|
1.923
|
NM_000427
|
LOR
|
loricrin
|
|
chr3_+_196295708
|
1.922
|
NM_001105573
|
FBXO45
|
F-box protein 45
|
|
chr17_-_46688362
|
1.916
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr15_+_38544966
|
1.910
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chrX_+_105066531
|
1.902
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr6_-_94129059
|
1.900
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr12_-_25055228
|
1.856
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr1_+_26737232
|
1.852
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr14_+_72398816
|
1.810
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr16_+_81348564
|
1.797
|
NM_022041
|
GAN
|
gigaxonin
|
|
chr17_-_9862765
|
1.773
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr11_-_133402227
|
1.773
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr11_-_132812870
|
1.770
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr16_-_3285359
|
1.730
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr12_-_25055966
|
1.714
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr11_-_797955
|
1.705
|
NM_001191060
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr14_-_88459469
|
1.689
|
NM_000153
NM_001201401
|
GALC
|
galactosylceramidase
|
|
chr17_-_9940004
|
1.653
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr11_-_798196
|
1.606
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr16_-_89007605
|
1.604
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr16_-_52580805
|
1.590
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr8_+_92082423
|
1.588
|
NM_016023
|
OTUD6B
|
OTU domain containing 6B
|
|
chrX_-_99665270
|
1.585
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr17_-_9929567
|
1.561
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr6_-_7313352
|
1.546
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr15_+_79724857
|
1.539
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr7_+_114055048
|
1.538
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr19_-_47975244
|
1.523
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr14_-_88460008
|
1.510
|
NM_001201402
|
GALC
|
galactosylceramidase
|
|
chr12_-_25102281
|
1.507
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr4_-_47840058
|
1.493
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr20_-_656822
|
1.484
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr16_-_3285069
|
1.482
|
NM_001145447
NM_003454
|
ZNF200
|
zinc finger protein 200
|
|
chr9_-_79520878
|
1.468
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr16_-_52581713
|
1.448
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr3_-_53079285
|
1.360
|
NM_001005159
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr22_+_39898248
|
1.347
|
NM_019008
|
SMCR7L
|
Smith-Magenis syndrome chromosome region, candidate 7-like
|
|
chr19_+_47104490
|
1.346
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr10_+_72238529
|
1.317
|
NM_014431
|
KIAA1274
|
KIAA1274
|
|
chr21_-_40033617
|
1.302
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr6_-_86352635
|
1.288
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr7_+_94023872
|
1.284
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chrX_-_24690740
|
1.279
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr3_-_53080038
|
1.265
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr7_-_27205148
|
1.265
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr9_-_72287097
|
1.248
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr20_+_30598244
|
1.240
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr14_+_90863302
|
1.233
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr3_+_133293256
|
1.212
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chrX_-_24665454
|
1.196
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr5_-_158526698
|
1.192
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr6_-_86352564
|
1.186
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr12_-_77459198
|
1.184
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr21_-_46221662
|
1.142
|
NM_001202489
NM_003343
NM_182688
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2
|
|
chr1_-_86622107
|
1.130
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chrX_+_49832214
|
1.128
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr2_+_189839078
|
1.104
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chrX_+_49687212
|
1.097
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr9_+_132934685
|
1.083
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chrX_-_134156488
|
1.076
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr14_+_72399785
|
1.064
|
NM_001204416
NM_001204417
NM_001204418
NM_001204419
NM_001204420
NM_001204421
NM_001204422
NM_004296
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr3_+_184097860
|
1.056
|
NM_003741
|
CHRD
|
chordin
|
|
chr16_-_89043215
|
1.049
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr9_+_132962871
|
1.045
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr17_+_8213555
|
1.027
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr3_+_133292404
|
1.010
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_+_5832778
|
1.010
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr9_+_115983807
|
1.006
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr11_-_74178593
|
0.994
|
NM_005472
|
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3
|
|
chr6_+_133562488
|
0.993
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr18_+_19749403
|
0.981
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr22_-_17602142
|
0.972
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr10_+_11206928
|
0.966
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_207224298
|
0.954
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr2_-_51259536
|
0.950
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr10_+_11059854
|
0.941
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr12_-_102872329
|
0.929
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr2_-_142889269
|
0.928
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr8_+_121137297
|
0.906
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr9_-_140196702
|
0.901
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr7_-_559479
|
0.892
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr12_+_65004191
|
0.890
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr8_+_1449531
|
0.882
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr10_+_68685791
|
0.859
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr17_+_53342320
|
0.848
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr18_-_24442534
|
0.843
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr11_+_7273141
|
0.820
|
NM_175733
|
SYT9
|
synaptotagmin IX
|
|
chr11_-_110583120
|
0.809
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr3_-_150264280
|
0.800
|
NM_014445
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr12_+_66218141
|
0.782
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr10_+_11047253
|
0.763
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_242641310
|
0.760
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr13_-_102568994
|
0.732
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chrX_+_47696281
|
0.723
|
NM_007137
|
ZNF81
|
zinc finger protein 81
|
|
chr17_-_47439326
|
0.713
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr17_+_68165660
|
0.712
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr2_-_50574891
|
0.690
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr9_+_110045516
|
0.687
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr16_-_12010518
|
0.686
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr5_+_148724976
|
0.678
|
NM_152407
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr12_+_50794591
|
0.663
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr17_+_27717942
|
0.656
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr1_+_41444968
|
0.645
|
NM_001905
|
CTPS
|
CTP synthase
|
|
chr6_-_86352982
|
0.635
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr4_+_106473776
|
0.626
|
NM_001242729
NM_017700
|
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38
|
|
chr1_+_193091057
|
0.624
|
NM_024529
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr1_-_115259433
|
0.623
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr6_+_4776641
|
0.622
|
NM_004824
|
CDYL
|
chromodomain protein, Y-like
|
|
chr17_-_46682333
|
0.619
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr19_-_51071279
|
0.611
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr1_+_18957499
|
0.607
|
NM_001135254
NM_002584
NM_013945
|
PAX7
|
paired box 7
|
|
chr6_+_24403144
|
0.593
|
NM_020662
|
MRS2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae)
|
|
chr16_-_12009804
|
0.586
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr5_-_178157597
|
0.581
|
NM_005649
|
ZNF354A
|
zinc finger protein 354A
|
|
chr21_-_39870303
|
0.571
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr4_+_113066552
|
0.569
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr14_+_55493837
|
0.557
|
NM_080867
NM_199421
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr18_-_24445652
|
0.557
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr16_+_69220940
|
0.547
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr1_-_53793743
|
0.541
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr5_+_56110899
|
0.535
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr2_+_113239733
|
0.531
|
NM_153712
|
TTL
|
tubulin tyrosine ligase
|
|
chr3_+_85008031
|
0.531
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr6_-_86351156
|
0.528
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr6_+_4836331
|
0.522
|
NM_001143970
|
CDYL
|
chromodomain protein, Y-like
|
|
chr1_+_60280462
|
0.506
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr3_+_85775631
|
0.504
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chrX_-_74145280
|
0.492
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr15_+_90544608
|
0.481
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr5_-_35230822
|
0.467
|
NM_000949
|
PRLR
|
prolactin receptor
|
|
chr4_+_88720701
|
0.450
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr18_+_55711388
|
0.448
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_57400199
|
0.447
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr14_-_57735614
|
0.431
|
NM_006544
|
EXOC5
|
exocyst complex component 5
|
|
chr2_+_63277191
|
0.430
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr1_-_17765052
|
0.426
|
NM_001136204
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr13_-_103054027
|
0.406
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr3_-_101039403
|
0.390
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr1_+_50513685
|
0.389
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_-_103574051
|
0.389
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr8_-_145559831
|
0.388
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr6_+_4890225
|
0.388
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chr21_-_28339405
|
0.386
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr22_+_39853316
|
0.386
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr12_-_102874373
|
0.382
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_17766241
|
0.382
|
NM_018715
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr17_-_71228482
|
0.378
|
NM_001098832
NM_032837
|
FAM104A
|
family with sequence similarity 104, member A
|
|
chr3_-_47823386
|
0.377
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr15_-_38856830
|
0.374
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr1_+_113615787
|
0.372
|
NM_014813
|
LRIG2
|
leucine-rich repeats and immunoglobulin-like domains 2
|
|
chr1_+_101003692
|
0.367
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr5_+_38845962
|
0.366
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr14_-_95608054
|
0.363
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr16_+_70380763
|
0.362
|
NM_018332
|
DDX19A
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19A
|
|
chr1_-_204380903
|
0.361
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr14_-_95623665
|
0.360
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr6_-_13814546
|
0.349
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr1_+_50574589
|
0.338
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr20_-_32700082
|
0.337
|
NM_003908
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|
|
chr20_+_37434294
|
0.335
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chrX_-_124097601
|
0.334
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr7_-_28220342
|
0.333
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr1_+_50571963
|
0.324
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr22_+_39883228
|
0.315
|
NM_001098270
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr10_-_108924287
|
0.310
|
NM_001013031
NM_001206569
NM_001206570
NM_001206571
NM_001206572
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr2_+_63277936
|
0.303
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chrX_-_24045302
|
0.302
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr12_+_27933162
|
0.300
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr9_-_95166827
|
0.283
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr10_+_98592658
|
0.278
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr19_+_14063318
|
0.274
|
NM_138353
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr8_-_87755880
|
0.274
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr12_-_70004941
|
0.272
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr1_-_205601999
|
0.261
|
NM_001973
NM_021795
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr1_+_50569581
|
0.257
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_+_101498028
|
0.251
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr8_+_98656329
|
0.250
|
NM_178812
|
MTDH
|
metadherin
|
|
chr21_-_30365111
|
0.246
|
NM_015565
|
LTN1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr1_+_4715104
|
0.241
|
NM_001042478
NM_018836
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr1_+_36023389
|
0.236
|
NM_001014839
NM_001014841
NM_014284
|
NCDN
|
neurochondrin
|
|
chr11_-_6502563
|
0.234
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr4_+_174089889
|
0.230
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr7_-_23509972
|
0.229
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr14_-_95599794
|
0.228
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr5_+_139493689
|
0.225
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|