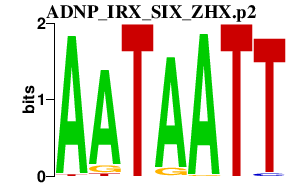
|
chr21_-_15918635
|
22.169
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr4_-_39033981
|
14.986
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr3_+_151488214
|
11.905
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr19_+_45116921
|
9.363
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr10_+_5136556
|
9.254
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_5136613
|
9.144
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_5136589
|
8.907
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr18_+_52258389
|
8.523
|
NM_173629
|
C18orf26
|
chromosome 18 open reading frame 26
|
|
chr15_-_76304762
|
8.486
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr10_+_5005685
|
7.797
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr2_-_224467049
|
7.610
|
|
SCG2
|
secretogranin II
|
|
chr4_+_41614911
|
7.526
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr16_+_82068830
|
6.997
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr16_+_82068910
|
6.919
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr2_+_11674241
|
6.872
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr16_+_82068958
|
6.778
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr17_-_39092714
|
6.340
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr20_+_58630965
|
6.336
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr12_+_101988746
|
6.209
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr4_+_41700736
|
6.093
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr10_+_5005601
|
5.888
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr2_+_29038862
|
5.836
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr4_+_41614725
|
5.803
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_-_224467095
|
5.697
|
|
SCG2
|
secretogranin II
|
|
chr17_-_67240955
|
5.661
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr2_-_224467142
|
5.404
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr13_+_36050885
|
5.169
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr10_+_70847857
|
4.932
|
|
SRGN
|
serglycin
|
|
chr2_+_234580504
|
4.922
|
NM_021027
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr19_+_49468557
|
4.885
|
NM_000146
|
FTL
|
ferritin, light polypeptide
|
|
chr19_+_49468587
|
4.856
|
|
FTL
|
ferritin, light polypeptide
|
|
chr12_+_86268072
|
4.734
|
NM_006183
|
NTS
|
neurotensin
|
|
chr10_-_5045967
|
4.728
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr8_-_108510094
|
4.596
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr4_+_41614933
|
4.568
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_70626429
|
4.484
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chrX_+_28605558
|
4.470
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr4_-_122148620
|
4.462
|
NM_001244764
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr2_-_99917638
|
4.415
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr1_+_61869699
|
4.404
|
|
NFIA
|
nuclear factor I/A
|
|
chr8_+_17396285
|
4.390
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr6_-_138189313
|
4.380
|
|
|
|
|
chr4_-_122085475
|
4.280
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chrX_-_15402469
|
4.061
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr10_+_5005453
|
3.967
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr19_+_49468656
|
3.961
|
|
FTL
|
ferritin, light polypeptide
|
|
chr2_-_190927321
|
3.738
|
NM_005259
|
MSTN
|
myostatin
|
|
chr10_+_114154522
|
3.681
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr19_-_39402797
|
3.634
|
NM_001243212
|
LOC643669
|
uncharacterized LOC643669
|
|
chr1_+_78470593
|
3.631
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr1_-_162346579
|
3.618
|
NM_182581
|
C1orf111
|
chromosome 1 open reading frame 111
|
|
chr16_+_86612114
|
3.528
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr12_-_91576579
|
3.514
|
|
DCN
|
decorin
|
|
chr5_-_42811959
|
3.472
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr1_+_201979689
|
3.470
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr11_+_10476665
|
3.437
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr7_-_107880492
|
3.390
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr12_-_91576805
|
3.308
|
NM_001920
|
DCN
|
decorin
|
|
chr1_+_201979747
|
3.306
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr17_-_202557
|
3.162
|
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr17_-_39093671
|
3.144
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_+_210444364
|
3.140
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr10_+_91061705
|
3.129
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr11_+_36317724
|
3.109
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr19_+_39899749
|
3.061
|
|
|
|
|
chr4_+_74301932
|
3.057
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr4_-_70725792
|
3.033
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr5_+_42756919
|
3.020
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr7_+_144015217
|
2.974
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr11_+_5710926
|
2.946
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr16_+_70207927
|
2.928
|
NM_173619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr17_-_202617
|
2.901
|
NM_001190411
NM_001190412
NM_001190413
NM_006987
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr1_+_168250277
|
2.836
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr7_-_27183225
|
2.826
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr10_+_70980058
|
2.689
|
NM_025130
|
HKDC1
|
hexokinase domain containing 1
|
|
chr6_-_30899926
|
2.679
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr4_+_3315873
|
2.622
|
NM_002926
NM_198229
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr1_+_18807423
|
2.616
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr19_-_48752921
|
2.580
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr2_+_234545099
|
2.562
|
NM_019075
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10
|
|
chr17_+_4692226
|
2.559
|
NM_001014985
|
GLTPD2
|
glycolipid transfer protein domain containing 2
|
|
chr2_+_169643048
|
2.541
|
NM_001171631
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr14_+_39703119
|
2.513
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr9_+_103340335
|
2.470
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr7_-_25268057
|
2.470
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr10_-_92681025
|
2.454
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr14_-_65409445
|
2.453
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr17_-_72855884
|
2.408
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr11_+_5710816
|
2.390
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr4_+_47033294
|
2.367
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr10_+_124320180
|
2.346
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_-_67138013
|
2.302
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr3_-_73673978
|
2.301
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr2_-_183387063
|
2.290
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr10_+_70847817
|
2.287
|
NM_002727
|
SRGN
|
serglycin
|
|
chr4_-_76944585
|
2.286
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr2_+_234637772
|
2.283
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr1_-_232651227
|
2.276
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr13_+_31309644
|
2.276
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr1_+_145524989
|
2.242
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr7_-_76829109
|
2.188
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr1_+_6106173
|
2.185
|
NM_001199863
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr5_-_1882764
|
2.179
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr17_-_15554809
|
2.176
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr6_+_88054570
|
2.155
|
NM_001010868
|
C6orf163
|
chromosome 6 open reading frame 163
|
|
chr18_+_55862577
|
2.135
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_+_36397534
|
2.125
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr7_-_80141241
|
2.114
|
NM_001102386
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3
|
|
chr22_-_38506674
|
2.098
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr1_+_6105980
|
2.092
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_+_112047088
|
2.088
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr18_+_55714457
|
2.078
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_+_234627423
|
2.066
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chrX_-_114797017
|
2.066
|
|
|
|
|
chr2_+_171571800
|
2.057
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr6_-_49712055
|
2.057
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr6_-_33041266
|
2.049
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr11_-_18258315
|
2.043
|
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr2_+_71162995
|
2.038
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chr2_-_192711923
|
2.034
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr5_-_95748688
|
2.015
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr2_+_234526290
|
2.007
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr4_-_2230957
|
1.959
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr13_+_31309690
|
1.941
|
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr7_-_27205075
|
1.937
|
|
HOXA9
|
homeobox A9
|
|
chr6_-_87804773
|
1.935
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr8_+_92261515
|
1.933
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr7_-_27205148
|
1.928
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr1_+_61547388
|
1.928
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_64088886
|
1.919
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr6_+_64286348
|
1.915
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr21_+_15588465
|
1.907
|
NM_144770
|
RBM11
|
RNA binding motif protein 11
|
|
chr3_+_151591428
|
1.899
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr12_+_60083125
|
1.893
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr12_+_8276227
|
1.890
|
NM_016184
NM_194447
NM_194448
NM_194450
|
CLEC4A
|
C-type lectin domain family 4, member A
|
|
chr19_-_42931500
|
1.887
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr2_+_211458078
|
1.887
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr1_-_204183036
|
1.884
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr7_-_7575449
|
1.870
|
NM_001037763
|
COL28A1
|
collagen, type XXVIII, alpha 1
|
|
chr19_-_18385166
|
1.854
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chrM_+_9725
|
1.846
|
|
|
|
|
chr10_+_99344421
|
1.845
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr1_+_61924621
|
1.833
|
|
NFIA
|
nuclear factor I/A
|
|
chrM_+_10361
|
1.818
|
|
|
|
|
chr12_+_104659046
|
1.816
|
|
RPL18A
|
ribosomal protein L18a
|
|
chr22_-_38506532
|
1.810
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr11_-_18062304
|
1.793
|
NM_004179
|
TPH1
|
tryptophan hydroxylase 1
|
|
chr3_-_101039403
|
1.775
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr2_-_188419049
|
1.764
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr3_-_39321479
|
1.753
|
NM_001337
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr10_-_128209996
|
1.739
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chrM_-_379
|
1.732
|
|
|
|
|
chr5_-_95158465
|
1.730
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr8_-_86290333
|
1.718
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr7_-_27205123
|
1.713
|
|
HOXA9
|
homeobox A9
|
|
chr17_+_7788094
|
1.712
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chrM_+_10704
|
1.708
|
|
|
|
|
chr1_-_108231122
|
1.706
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_+_145525041
|
1.701
|
|
ITGA10
|
integrin, alpha 10
|
|
chr1_-_54483802
|
1.682
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr11_+_120973374
|
1.680
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr2_+_33738941
|
1.680
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr11_+_87032428
|
1.675
|
|
TMEM135
|
transmembrane protein 135
|
|
chr22_+_22676814
|
1.675
|
|
|
|
|
chr14_-_92198383
|
1.658
|
NM_024764
|
CATSPERB
|
cation channel, sperm-associated, beta
|
|
chr5_-_149324282
|
1.657
|
NM_000440
|
PDE6A
|
phosphodiesterase 6A, cGMP-specific, rod, alpha
|
|
chr4_-_23891608
|
1.644
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr5_-_95158365
|
1.633
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr2_-_192711651
|
1.633
|
|
SDPR
|
serum deprivation response
|
|
chr5_+_140797210
|
1.629
|
NM_032101
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr18_-_45663667
|
1.623
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr12_-_16757944
|
1.573
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_+_102965836
|
1.572
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chr4_-_149357954
|
1.560
|
|
|
|
|
chr17_+_7741630
|
1.555
|
|
|
|
|
chr5_-_59783885
|
1.548
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_109605668
|
1.541
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr17_+_34087915
|
1.531
|
NM_145272
|
C17orf50
|
chromosome 17 open reading frame 50
|
|
chr1_-_217250230
|
1.529
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr9_+_120466625
|
1.526
|
|
TLR4
|
toll-like receptor 4
|
|
chr3_-_187452669
|
1.521
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_+_169926084
|
1.513
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr5_+_67586466
|
1.513
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr4_+_78432905
|
1.491
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr2_+_171571936
|
1.476
|
|
|
|
|
chr13_-_30424795
|
1.476
|
NM_007106
|
UBL3
|
ubiquitin-like 3
|
|
chr15_-_56209305
|
1.469
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr9_+_109686436
|
1.463
|
|
ZNF462
|
zinc finger protein 462
|
|
chrM_-_7189
|
1.456
|
|
|
|
|
chr1_-_161279724
|
1.434
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr5_-_58882274
|
1.432
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_87770267
|
1.418
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr15_+_34261088
|
1.412
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr9_-_95186559
|
1.409
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr1_+_79086087
|
1.404
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr6_-_132272311
|
1.401
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chrX_+_70765479
|
1.400
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr2_-_200325200
|
1.393
|
|
SATB2
|
SATB homeobox 2
|
|
chr20_-_34195409
|
1.389
|
|
FER1L4
|
fer-1-like 4 (C. elegans) pseudogene
|
|
chr2_-_200329810
|
1.386
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|