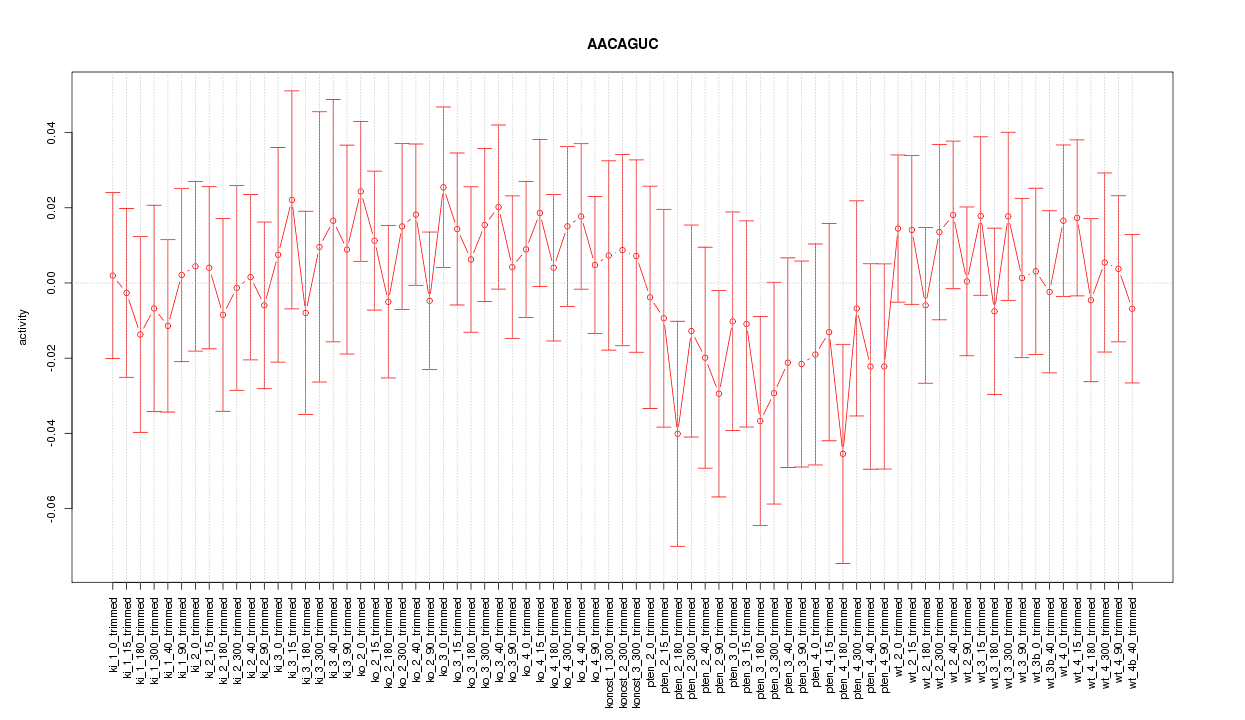
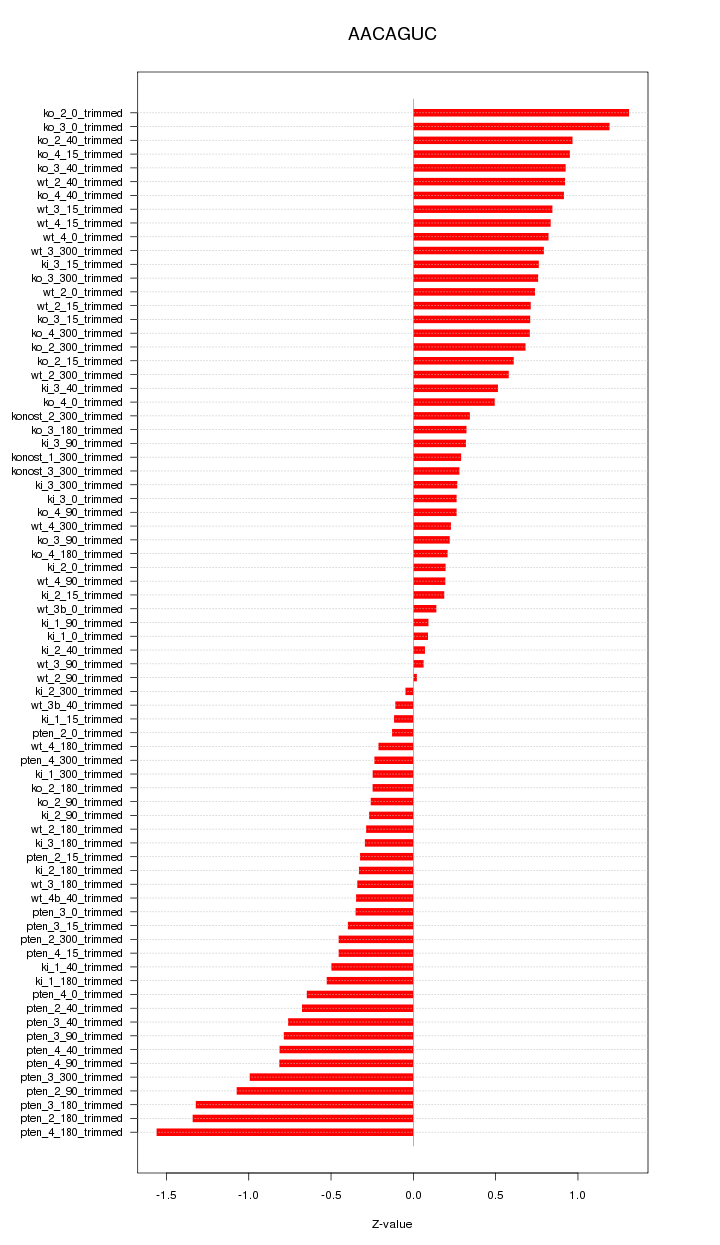
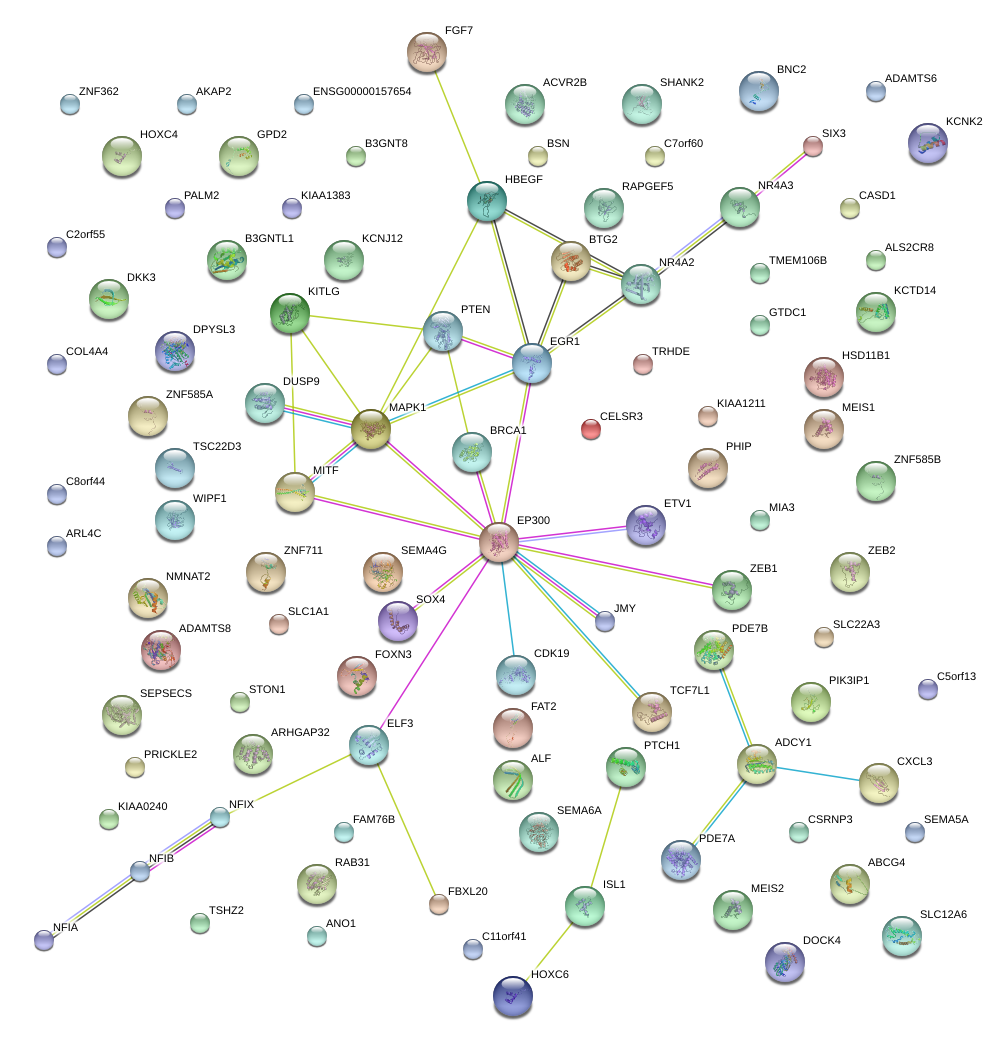
|
chr3_-_64211111
|
5.154
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr10_+_102732285
|
3.953
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr5_-_111093251
|
3.681
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_115910406
|
3.538
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr11_+_69924407
|
3.513
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr5_-_111312522
|
3.318
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
3.138
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
3.083
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093030
|
2.993
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr19_-_37663567
|
2.741
|
NM_152655
|
ZNF585A
|
zinc finger protein 585A
|
|
chr19_-_37701385
|
2.540
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr1_-_183387501
|
2.494
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr5_+_137801178
|
2.358
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr11_-_12030128
|
2.336
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr9_+_112403067
|
2.307
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr1_+_215256559
|
2.204
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr1_-_183274008
|
2.153
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr17_+_21279667
|
2.142
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr22_-_31688329
|
1.875
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr5_-_111091947
|
1.805
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_228029274
|
1.767
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr3_-_48700317
|
1.713
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr19_-_37663332
|
1.711
|
NM_199126
|
ZNF585A
|
zinc finger protein 585A
|
|
chr6_+_136172802
|
1.699
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_145277879
|
1.669
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_77734285
|
1.629
|
NM_023930
|
KCTD14
|
potassium channel tetramerisation domain containing 14
|
|
chr2_+_48757292
|
1.589
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr3_+_38495778
|
1.555
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr5_-_146833150
|
1.491
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr11_-_12030882
|
1.485
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr5_-_64777703
|
1.449
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr12_+_72666462
|
1.423
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr6_-_111136407
|
1.422
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr1_+_209878135
|
1.397
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr18_+_9708187
|
1.395
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr11_-_12030622
|
1.379
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr10_+_89623091
|
1.308
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr4_+_57036360
|
1.255
|
NM_020722
|
KIAA1211
|
KIAA1211
|
|
chrX_+_84498993
|
1.252
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr9_+_4490193
|
1.223
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr17_+_21308447
|
1.141
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chr22_-_22221953
|
1.129
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr5_-_146889618
|
1.091
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr11_-_70507911
|
1.071
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_+_21593963
|
1.031
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr1_+_209859524
|
1.019
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr4_-_74904329
|
1.006
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chrX_+_152907892
|
0.981
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr11_-_70935807
|
0.962
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_-_99552677
|
0.956
|
NM_207362
|
C2orf55
|
chromosome 2 open reading frame 55
|
|
chr9_-_16870719
|
0.930
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr20_+_51801821
|
0.923
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr17_-_37557851
|
0.914
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr6_+_160769365
|
0.906
|
NM_021977
|
SLC22A3
|
solute carrier family 22 (extraneuronal monoamine transporter), member 3
|
|
chr14_-_90085325
|
0.876
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr1_+_33722158
|
0.872
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr2_+_157292826
|
0.845
|
NM_000408
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
|
|
chr14_-_89883306
|
0.809
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr9_-_14398981
|
0.783
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr19_-_41934634
|
0.781
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr20_+_51588945
|
0.763
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr7_-_14025826
|
0.752
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr2_+_157291951
|
0.743
|
NM_001083112
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
|
|
chr9_-_98279246
|
0.702
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr9_-_14314036
|
0.696
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr5_-_150948504
|
0.686
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr5_-_139726173
|
0.683
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr1_+_215178884
|
0.682
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr8_-_66753968
|
0.667
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr9_-_98269480
|
0.652
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr5_+_78531848
|
0.650
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr7_+_12250818
|
0.641
|
NM_001134232
NM_018374
|
TMEM106B
|
transmembrane protein 106B
|
|
chr2_+_45169036
|
0.638
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chrX_-_106959597
|
0.631
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_+_69788562
|
0.624
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_222791151
|
0.613
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr1_+_203274646
|
0.608
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr7_-_22396511
|
0.596
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr7_-_111846384
|
0.588
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr3_+_69915374
|
0.577
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_-_98270321
|
0.567
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr2_+_203776930
|
0.566
|
NM_001104586
NM_024744
|
ALS2CR8
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8
|
|
chrX_-_106960268
|
0.563
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_-_88974094
|
0.562
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr5_+_50678957
|
0.561
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr11_+_119020298
|
0.546
|
NM_001142505
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr2_-_175547471
|
0.544
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr3_+_69812620
|
0.542
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_49591897
|
0.542
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr2_-_235405692
|
0.536
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr17_-_41277409
|
0.527
|
NM_007297
NM_007299
NM_007294
NM_007300
|
BRCA1
|
breast cancer 1, early onset
|
|
chr6_+_42788793
|
0.524
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr3_+_69985750
|
0.521
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69812961
|
0.516
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_+_49715374
|
0.508
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr2_+_166428845
|
0.504
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr11_-_95522945
|
0.490
|
NM_144664
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr9_-_98269686
|
0.487
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr17_-_41276112
|
0.485
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chr15_-_34630203
|
0.477
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr7_+_94138991
|
0.473
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr8_+_67579786
|
0.473
|
NM_001204173
NM_019607
|
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough
chromosome 8 open reading frame 44
|
|
chr2_+_85360373
|
0.470
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr15_-_34629941
|
0.468
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_+_66662516
|
0.468
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr7_-_112579726
|
0.462
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr7_-_14030864
|
0.461
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chrX_-_107019001
|
0.460
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr8_-_66701175
|
0.454
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr2_-_157189040
|
0.453
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr5_-_9546157
|
0.453
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr7_+_45614081
|
0.452
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr6_-_79787939
|
0.451
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr11_+_33563772
|
0.450
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr2_-_145090038
|
0.448
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr1_+_232940637
|
0.446
|
NM_019090
|
KIAA1383
|
KIAA1383
|
|
chr1_+_201979689
|
0.445
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr2_+_166326156
|
0.443
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr2_-_175499275
|
0.431
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_-_25161908
|
0.429
|
NM_016955
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase
|
|
chr11_+_119019749
|
0.425
|
NM_022169
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr15_-_34629044
|
0.419
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr22_+_41488607
|
0.418
|
NM_001429
|
EP300
|
E1A binding protein p300
|
|
chr12_+_54447660
|
0.415
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr11_-_128894008
|
0.415
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr7_+_18548899
|
0.411
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr9_-_98270830
|
0.407
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr7_-_100493481
|
0.404
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr17_-_43045612
|
0.397
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr2_+_5832778
|
0.382
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr14_+_59655351
|
0.369
|
NM_014992
|
DAAM1
|
dishevelled associated activator of morphogenesis 1
|
|
chr8_+_67687415
|
0.363
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr4_+_124320664
|
0.356
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_1795004
|
0.355
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr9_-_3525973
|
0.345
|
NM_002919
NM_134428
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr11_-_16430425
|
0.342
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_+_16964730
|
0.341
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr7_+_18126543
|
0.341
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr6_+_87865261
|
0.337
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr17_+_47865980
|
0.337
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr8_+_67624652
|
0.320
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr6_+_49431090
|
0.317
|
NM_018132
|
CENPQ
|
centromere protein Q
|
|
chr1_+_107682628
|
0.316
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr4_+_144257982
|
0.315
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr1_-_184006828
|
0.314
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr3_+_43732358
|
0.310
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr8_+_124780742
|
0.305
|
NM_144963
|
FAM91A1
|
family with sequence similarity 91, member A1
|
|
chr3_-_168864325
|
0.302
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr7_-_14029641
|
0.301
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr12_+_51632507
|
0.299
|
NM_001136264
NM_001136266
NM_001136267
NM_001136268
NM_001136269
NM_014764
|
DAZAP2
|
DAZ associated protein 2
|
|
chr2_-_110962577
|
0.298
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr3_-_15563160
|
0.297
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr7_+_105603656
|
0.294
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr15_-_60884615
|
0.290
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_-_169863048
|
0.289
|
NM_020423
NM_181093
|
SCYL3
|
SCY1-like 3 (S. cerevisiae)
|
|
chr9_+_72658496
|
0.288
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr1_-_43424834
|
0.283
|
NM_006516
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr13_+_42846255
|
0.281
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr16_+_19566736
|
0.279
|
NM_020314
|
C16orf62
|
chromosome 16 open reading frame 62
|
|
chr6_+_34433849
|
0.278
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr6_-_132272311
|
0.278
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr6_-_119399759
|
0.277
|
NM_024581
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr16_+_29823408
|
0.275
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr1_-_160232286
|
0.275
|
NM_015726
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr3_+_180630054
|
0.273
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr3_-_15540378
|
0.269
|
NM_080538
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr10_+_114709963
|
0.265
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr5_+_143550395
|
0.262
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr2_-_120980971
|
0.258
|
NM_024121
|
TMEM185B
|
transmembrane protein 185B
|
|
chr6_+_130339715
|
0.255
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr3_+_186501360
|
0.254
|
NM_001967
|
EIF4A2
|
eukaryotic translation initiation factor 4A2
|
|
chr7_-_14031049
|
0.252
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr7_-_105319608
|
0.246
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr13_+_53029494
|
0.244
|
NM_001098525
NM_018204
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr5_-_171433876
|
0.244
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr8_-_81787015
|
0.243
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr1_-_156051788
|
0.242
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr11_-_16497917
|
0.240
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr9_+_86595636
|
0.240
|
NM_024945
|
RMI1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae)
|
|
chr8_+_97505876
|
0.238
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr18_+_55102777
|
0.238
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr12_-_76478737
|
0.236
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr2_+_166095911
|
0.235
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr11_+_111789600
|
0.233
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr8_+_17013799
|
0.232
|
NM_016353
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr12_-_3982394
|
0.231
|
NM_020367
|
PARP11
|
poly (ADP-ribose) polymerase family, member 11
|
|
chrX_+_122993536
|
0.228
|
NM_001204401
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr2_+_32390902
|
0.227
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr6_+_34482646
|
0.224
|
NM_001199583
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr15_-_34610928
|
0.223
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr4_+_124317884
|
0.222
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr6_-_139695349
|
0.221
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr13_+_37393306
|
0.221
|
NM_000538
|
RFXAP
|
regulatory factor X-associated protein
|
|
chr1_+_36348795
|
0.220
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chrX_-_50557019
|
0.217
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr14_-_73493744
|
0.216
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr8_+_15397595
|
0.210
|
NM_006765
NM_178234
|
TUSC3
|
tumor suppressor candidate 3
|
|
chr3_+_178866124
|
0.209
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr7_-_91875296
|
0.209
|
NM_194455
NM_194456
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr12_+_107168336
|
0.207
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|