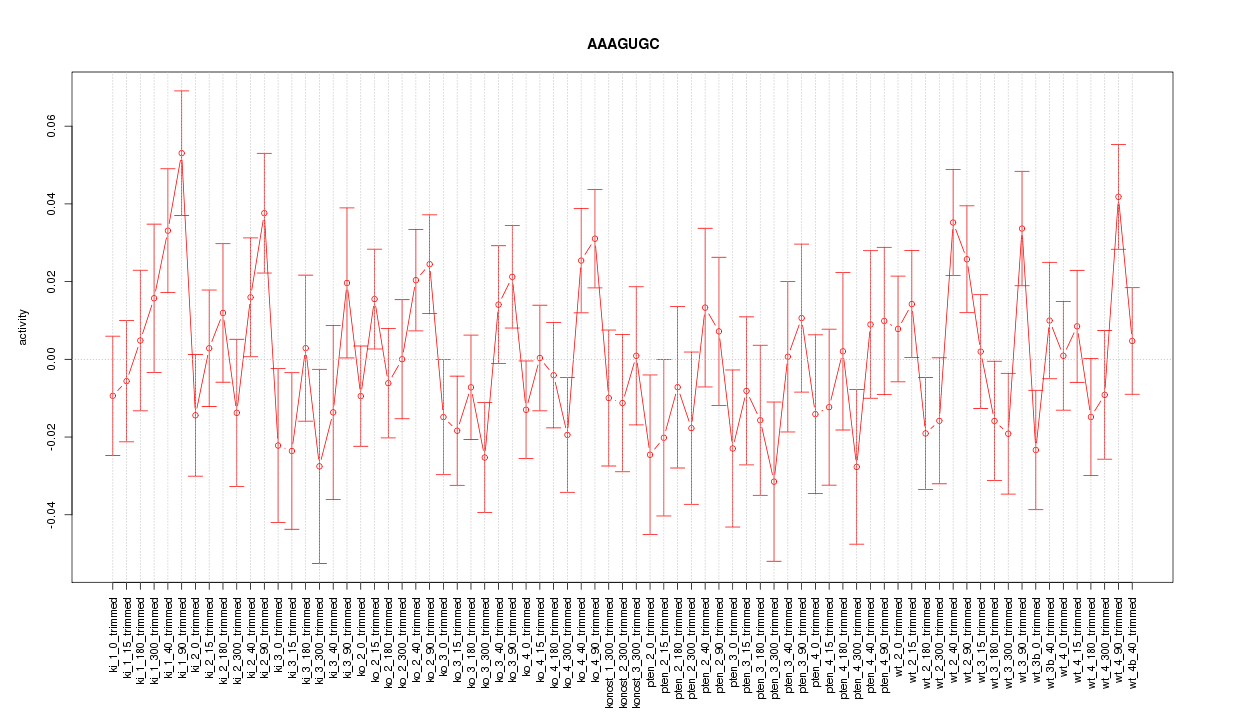
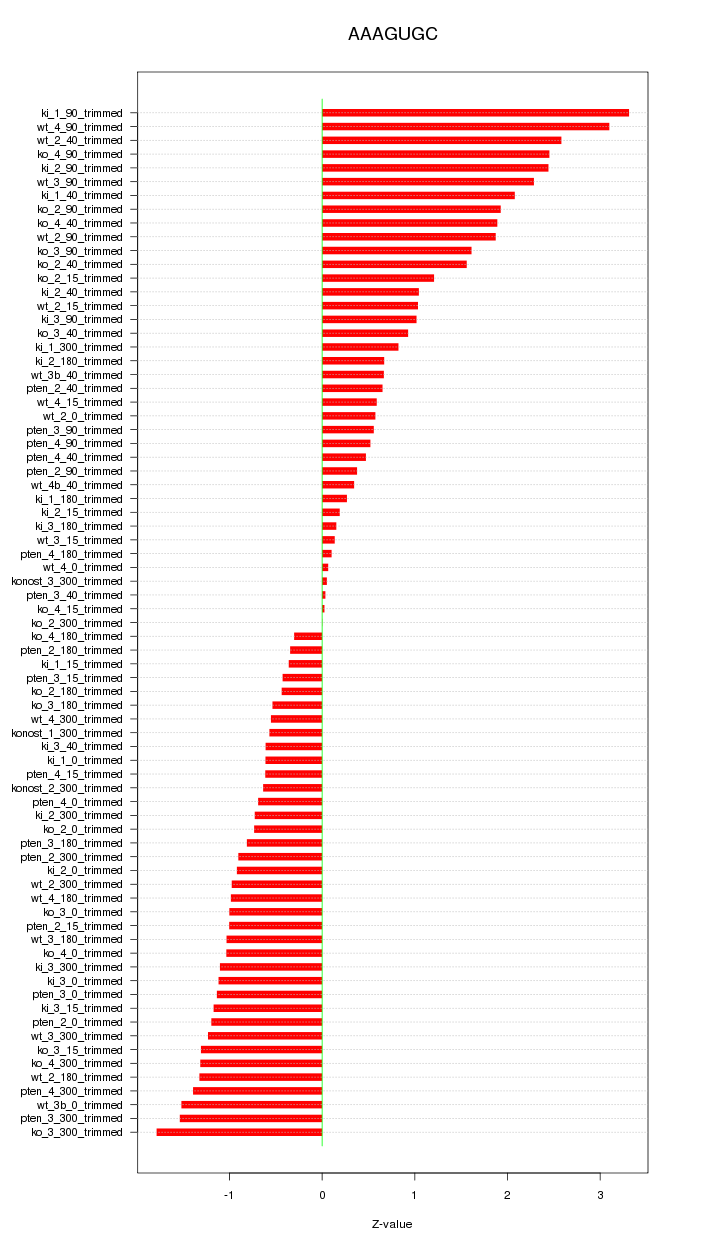
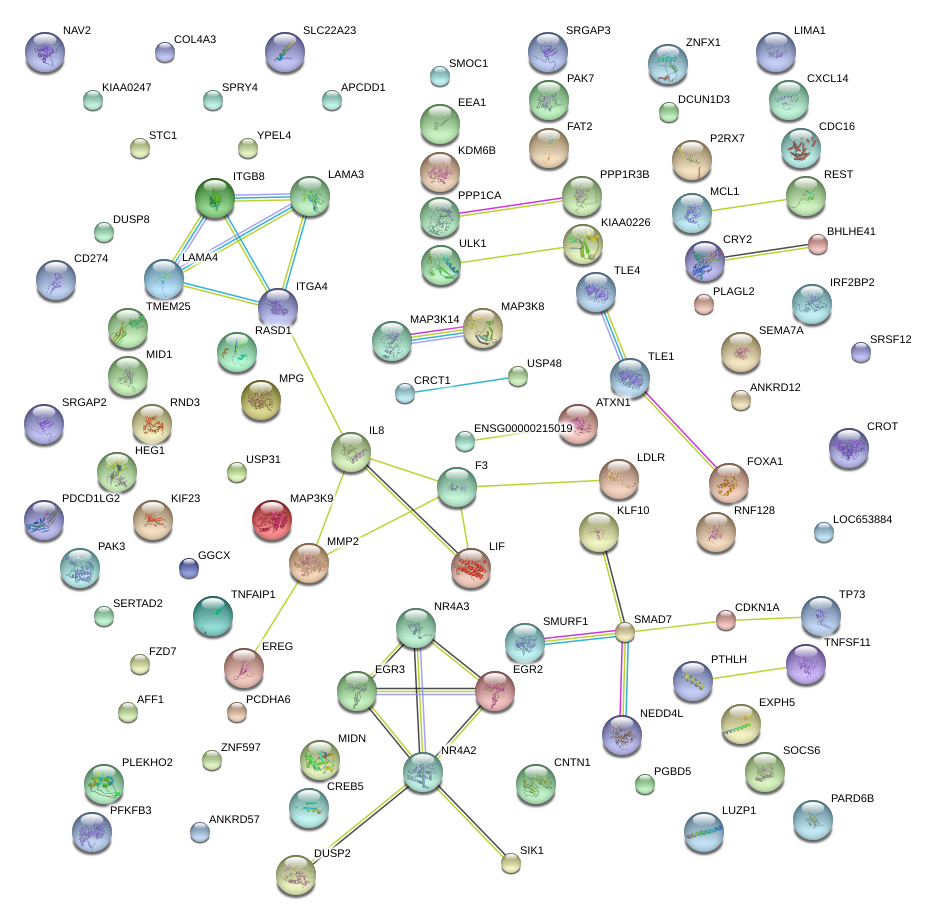
|
chr8_-_22550057
|
8.445
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
8.391
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
8.137
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr2_-_96811138
|
7.847
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr9_+_102584136
|
7.099
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr15_-_74726258
|
6.671
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr10_-_64576123
|
6.666
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_64578926
|
6.537
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr22_-_30642683
|
5.817
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr4_+_74606222
|
5.639
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr15_-_74726000
|
5.396
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr9_+_5450455
|
5.315
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr14_-_38064440
|
5.231
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr6_-_89827605
|
4.882
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr17_+_7743233
|
4.594
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr19_+_1248548
|
4.484
|
NM_177401
|
MIDN
|
midnolin
|
|
chr17_-_43394326
|
4.396
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr18_+_10454620
|
4.258
|
NM_153000
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr1_-_95007139
|
4.218
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr8_-_9009050
|
4.137
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr8_-_9008138
|
4.118
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr5_-_134914968
|
3.953
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr20_+_49348050
|
3.672
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr5_-_141704575
|
3.652
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr16_+_55512801
|
3.486
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_+_152486977
|
3.390
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr19_+_11200037
|
3.275
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr4_+_75230856
|
3.255
|
NM_001432
|
EREG
|
epiregulin
|
|
chr2_+_110371887
|
3.243
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr20_-_30795346
|
3.204
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr1_-_230513390
|
3.200
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr7_-_106301329
|
3.080
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr7_+_28452143
|
2.855
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_20370724
|
2.833
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr7_+_28338939
|
2.823
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_28475233
|
2.741
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_+_118401755
|
2.702
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr7_+_28725597
|
2.694
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_+_182321618
|
2.691
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr15_+_65134081
|
2.604
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr12_-_28122884
|
2.596
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr18_+_21269528
|
2.585
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr9_+_5510544
|
2.545
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr5_+_140254930
|
2.526
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr14_-_71275732
|
2.515
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr1_-_150552127
|
2.460
|
NM_001197320
NM_021960
NM_182763
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr6_+_36646455
|
2.396
|
NM_000389
NM_001220778
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr13_+_43148182
|
2.395
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr10_+_6186758
|
2.385
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr11_+_19734821
|
2.330
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr16_-_20911560
|
2.316
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr6_+_36644236
|
2.307
|
NM_001220777
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr11_-_1593149
|
2.292
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr10_+_6244839
|
2.290
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr1_-_23495347
|
2.273
|
NM_001142546
NM_033631
|
LUZP1
|
leucine zipper protein 1
|
|
chr11_-_57417247
|
2.258
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr3_-_124774735
|
2.220
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr2_-_85788644
|
2.216
|
NM_000821
NM_001142269
|
GGCX
|
gamma-glutamyl carboxylase
|
|
chr1_-_234745223
|
2.204
|
NM_001077397
NM_182972
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr17_-_17399494
|
2.198
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr21_-_44846927
|
2.197
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chrX_+_105969875
|
2.189
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr11_+_19372270
|
2.177
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr2_-_151344145
|
2.116
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr9_+_82186846
|
2.109
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr8_-_103667882
|
2.079
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr3_-_197476567
|
2.050
|
NM_001145642
|
KIAA0226
|
KIAA0226
|
|
chr12_-_26277807
|
2.043
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr12_-_28124915
|
2.028
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr18_+_21452981
|
2.026
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr8_-_23712297
|
2.004
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr1_+_3607232
|
1.997
|
NM_001126240
NM_001126241
NM_001126242
NM_001204189
NM_001204190
NM_001204191
|
TP73
|
tumor protein p73
|
|
chrX_-_10588458
|
1.982
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr18_+_55714457
|
1.959
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_157189040
|
1.947
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr20_-_9819406
|
1.866
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr3_-_197463713
|
1.856
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr18_+_55888750
|
1.850
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr8_-_103666018
|
1.830
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr14_+_70346113
|
1.806
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chrX_+_105937067
|
1.769
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr10_+_30722856
|
1.762
|
NM_001244134
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr2_-_64881040
|
1.751
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr16_+_55515468
|
1.748
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr12_-_50677238
|
1.740
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr18_+_55816546
|
1.734
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_-_10645726
|
1.720
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr16_-_23160509
|
1.704
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr15_+_69706619
|
1.694
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr1_+_3569098
|
1.688
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr4_+_87856152
|
1.670
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr3_-_9291251
|
1.662
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr6_-_3445198
|
1.661
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr18_+_55862577
|
1.659
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_67956136
|
1.613
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr18_+_55711388
|
1.588
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_-_3493489
|
1.576
|
NM_152457
|
ZNF597
|
zinc finger protein 597
|
|
chr12_+_132379262
|
1.541
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr14_+_70078309
|
1.540
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr7_+_86974928
|
1.536
|
NM_001143935
NM_001243745
NM_021151
|
CROT
|
carnitine O-octanoyltransferase
|
|
chr2_+_202899309
|
1.521
|
NM_003507
|
FZD7
|
frizzled family receptor 7
|
|
chr11_+_45868668
|
1.509
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr6_-_16761600
|
1.502
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr4_+_57774035
|
1.488
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chrX_-_10544852
|
1.472
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr11_+_45868956
|
1.467
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr6_-_3456734
|
1.459
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr18_-_46469118
|
1.416
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr17_+_26662547
|
1.403
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr11_-_108464344
|
1.399
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr12_-_93323088
|
1.399
|
NM_003566
|
EEA1
|
early endosome antigen 1
|
|
chr4_+_87928083
|
1.397
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr2_+_228029280
|
1.388
|
NM_000091
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen)
|
|
chr20_-_47894591
|
1.385
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr18_+_9137560
|
1.383
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr7_-_98741681
|
1.375
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr18_+_9136742
|
1.364
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr7_-_142583260
|
1.364
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chr11_+_121322848
|
1.358
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_-_38325224
|
1.342
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr9_-_14398981
|
1.322
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr10_-_30348433
|
1.305
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr13_+_93879011
|
1.285
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr12_-_53893195
|
1.280
|
NM_001193511
NM_006301
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12
|
|
chrX_+_16964730
|
1.279
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr13_+_43136871
|
1.269
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr12_-_50616425
|
1.250
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr18_-_46477080
|
1.248
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr6_-_152957985
|
1.238
|
NM_033071
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr4_+_57775064
|
1.217
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr6_-_47277646
|
1.214
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr1_-_204380903
|
1.209
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr5_+_140235594
|
1.198
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr10_+_1102741
|
1.191
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr18_+_48086402
|
1.185
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr16_+_9185534
|
1.178
|
NM_014117
|
C16orf72
|
chromosome 16 open reading frame 72
|
|
chr3_+_140950671
|
1.161
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr7_-_112430408
|
1.150
|
NM_022484
|
TMEM168
|
transmembrane protein 168
|
|
chr12_-_120315074
|
1.143
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr5_+_76011780
|
1.141
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr6_-_152958533
|
1.140
|
NM_182961
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_-_14314036
|
1.128
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr2_+_27193448
|
1.121
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr13_-_107187312
|
1.115
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr10_-_61469322
|
1.112
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr10_+_97803064
|
1.111
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr4_-_141677470
|
1.105
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr10_+_112404104
|
1.104
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr6_+_158244281
|
1.100
|
NM_016224
|
SNX9
|
sorting nexin 9
|
|
chr7_-_127032717
|
1.048
|
NM_176814
|
ZNF800
|
zinc finger protein 800
|
|
chr2_-_19558371
|
1.026
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr5_-_9546157
|
1.009
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr14_-_73360792
|
1.008
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr3_-_56809594
|
0.996
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr10_-_61122219
|
0.977
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr7_-_44924946
|
0.977
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr5_+_140165875
|
0.968
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr1_+_145438437
|
0.962
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr11_+_109964086
|
0.957
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr15_+_75287829
|
0.952
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr5_-_150948504
|
0.946
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr4_+_7045142
|
0.939
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr4_+_48343502
|
0.933
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr8_-_116681103
|
0.929
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr19_-_4066739
|
0.928
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr15_+_68871285
|
0.917
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chrX_-_10851808
|
0.913
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_-_179198814
|
0.911
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr20_+_1875382
|
0.899
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr1_+_183605207
|
0.893
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr16_+_85061303
|
0.889
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr17_-_62502410
|
0.886
|
NM_004396
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr14_-_45431121
|
0.884
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr15_+_68908878
|
0.880
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr20_+_1874779
|
0.877
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr5_+_140247767
|
0.875
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr18_-_46475702
|
0.874
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr11_-_18655962
|
0.863
|
NM_194285
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr2_+_169312793
|
0.859
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr11_-_123612324
|
0.859
|
NM_003455
|
ZNF202
|
zinc finger protein 202
|
|
chr4_+_88928797
|
0.858
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr3_-_57113292
|
0.858
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr20_+_1875825
|
0.857
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr5_+_140207562
|
0.856
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr5_+_140180782
|
0.845
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr10_+_120789227
|
0.844
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr2_+_74273449
|
0.837
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr3_-_46037299
|
0.830
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr5_+_172068188
|
0.822
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr16_-_67450149
|
0.819
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr1_+_160336856
|
0.809
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr4_+_120133764
|
0.798
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr6_+_33378772
|
0.794
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chr2_-_145052059
|
0.783
|
NM_001006636
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr2_-_204399906
|
0.782
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr5_+_140201221
|
0.778
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr11_-_8680382
|
0.776
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chrX_+_68048792
|
0.775
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr14_-_91526912
|
0.771
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr9_-_95527002
|
0.763
|
NM_001003800
NM_015250
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|